guide:7a0c7996ab: Difference between revisions
No edit summary |
No edit summary |
||
| Line 1: | Line 1: | ||
<div class="d-none"><math> | |||
\newcommand{\varitem}[1]{\vspace{0.1cm}\hspace*{1cm}\raisebox{0.1cm}{ \tikz[baseline=(SebastianoItem.base),remember | |||
picture]{% | |||
\node[fill=green!20,inner sep=4pt,font=\sffamily] (SebastianoItem){#1};}\hspace{0.25cm} \vspace*{0.1cm}}} | |||
\newcommand{\SebastianoHighlight}{\tikz[overlay,remember picture]{% | |||
\fill[red!20] ([yshift=-20pt,xshift=-\pgflinewidth]SebastianoItem.east) -- ++(2pt,-2pt) | |||
-- ++(-2pt,-2pt) -- cycle; | |||
}} | |||
\newcommand{\rz}[1]{\displaystyle\stackrel{#1}{\phantom{.}}} % to raise exponent just a bit | |||
\newcommand{\CC}{C\nolinebreak\hspace{-.05em}\raisebox{.4ex}{\tiny\bf +}\nolinebreak\hspace{-.10em}\raisebox{.4ex}{\tiny\bf +}} | |||
\newcommand{\bm}[1]{\boldsymbol{#1}} | |||
\newcommand{\sectionbreak}{\clearpage} | |||
\newcommand\pythonstyle{\lstset{ | |||
language=Python, | |||
basicstyle=\ttm, | |||
otherkeywords={self}, % Add keywords here | |||
keywordstyle=\ttb\color{deepblue}, | |||
emph={MyClass,__init__}, % Custom highlighting | |||
emphstyle=\ttb\color{deepred}, % Custom highlighting style | |||
stringstyle=\color{deepgreen}, | |||
frame=tb, % Any extra options here | |||
showstringspaces=false % | |||
}} | |||
\newcommand\pythoninline[1]{{\pythonstyle\lstinline!#1!}} | |||
\DeclareMathOperator{\sign}{sign} | |||
\newcommand{\mathds}{\mathbb}</math></div> | |||
\label{sec:unsupervised} | |||
In Sec.~[[guide:4b07d6b69a#sec:supervised |sec:supervised]], we discussed supervised learning tasks, for which datasets consist of input-output pairs, or data-label pairs. More often than not, however, we have data without labels and would like to extract information from such a dataset. Clustering problems fall in this category, for instance: We suspect that the data can be divided into different types, but we do not know which features distinguish these types. | |||
Mathematically, we can think of the data <math>\bm{x}</math> as samples that were drawn from a probability distribution <math>P(\bm{x})</math>. The unsupervised learning task is to implicitly represent this distribution with a model, for example represented by a neural network. The model can then be used to study properties of the distribution or to generate new ‘artificial’ data. The models we encounter in this chapter are thus also referred to as ''generative models''. In general, unsupervised learning is conceptually more challenging than supervised learning. At the same time, unsupervised algorithms are highly desirable, since unlabelled data is much more abundant than labelled data. Moreover, we can in principle use a generative model for a classification task by learning the joint probability distribution of the data-label pair. | |||
In this chapter, we will introduce three types of neural networks that are specific to unsupervised learning tasks: ''Restricted Boltzmann machines'', ''autoencoders'', and ''generative adversarial networks''. Furthermore, we will discuss how the RNN introduced in the previous chapter can also be used for an unsupervised task. | |||
===Restricted Boltzmann machine=== | |||
''Restricted Boltzmann Machines'' (RBM) are a class of generative stochastic neural networks. More specifically, given some (binary) input data <math>\bm{x}\in\{0,1\}^{n_v}</math>, an RBM can be trained to approximate the probability distribution of this input. Moreover, once the neural network is trained to approximate the distribution of the input, we can sample from the network, in other words we generate new instances from the learned probability distribution. | |||
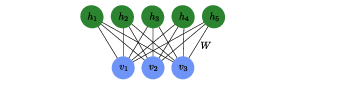
The RBM consists of two layers (see Fig.~[[#fig:RBM |fig:RBM]]) of ''binary units''. Each binary unit is a variable which can take the values <math>0</math> or <math>1</math>. | |||
We call the first (input) layer visible and the second layer hidden. | |||
The visible layer with input variables <math>\lbrace v_{1}, v_{2}, \dots v_{n_{\mathrm{v}}}\rbrace </math>, which we collect in the vector <math>\bm{v}</math>, is connected to the hidden layer with variables <math>\{ h_{1}, h_{2}, \dots h_{n_{\mathrm{h}}}\}</math>, which we collect in the vector <math>\bm{h}</math>. | |||
The role of the hidden layer is to mediate correlations between the units of the visible layer. | |||
In contrast to the neural networks we have seen in the previous chapter, the hidden layer is not followed by an output layer. Instead, the RBM represents a probability distribution <math>P_{\text{rbm}}(\bm{v})</math>, which depends on variational parameters represented by the weights and biases of a neural network. | |||
The RBM, as illustrated by the graph in Fig.~[[#fig:RBM |fig:RBM]], is a special case of a network structure known as a Boltzmann machine with the restriction that a unit in the visible layer is only connected to hidden units and vice versa, hence the name ''restricted'' Boltzmann machine. | |||
<div id="fig:RBM" class="d-flex justify-content-center"> | |||
[[File:guide_7132c_rbm.png | 400px | thumb | ''' Restricted Boltzmann machine.''' Each of the three visible units and five hidden units represents a variable that can take the values <math>\pm1</math> and the connections between them represent the entries <math>W_{ij}</math> of the weight matrix that enters the energy function~\eqref{eqn: RBM Energy}. ]] | |||
</div> | |||
The structure of the RBM is motivated from statistical physics: | |||
To each choice of the binary vectors <math>\bm{v}</math> and <math>\bm{h}</math>, we assign a value we call the energy | |||
<math display="block"> | |||
\begin{equation}\label{eqn: RBM Energy} | |||
E(\bm{v},\bm{h}) = -\sum_{i}a_{i}v_{i} - \sum_{j}b_{j}h_{j} - \sum_{ij} v_{i}W_{ij}h_{j}, | |||
\end{equation} | |||
</math> | |||
where the vectors <math>\bm{a}</math>, <math>\bm{b}</math>, and the matrix <math>W</math> are the variational parameters of the model. Given the energy, the probability distribution over the configurations <math>(\bm{v}, \bm{h})</math> is defined as | |||
<math display="block"> | |||
\begin{equation}\label{eqn: RBM Joint Probability} | |||
P_{\textrm{rbm}}(\bm{v},\bm{h}) = \frac{1}{Z}e^{-E(\bm{v},\bm{h})}, | |||
\end{equation} | |||
</math> | |||
where | |||
<math display="block"> | |||
\begin{equation} | |||
Z = \sum_{\bm{v},\bm{h}} e^{-E(\bm{v},\bm{h})} | |||
\label{eq: partition function} | |||
\end{equation} | |||
</math> | |||
is a normalisation factor called the partition function. | |||
The sum in Eq.~\eqref{eq: partition function} runs over all binary vectors <math>\bm{v}</math> and <math>\bm{h}</math>, i.e., vectors with entries <math>0</math> or <math>1</math>. | |||
The probability that the model assigns to a visible vector <math>\bm{v}</math> is then the marginal over the joint probability distribution Eq.~\eqref{eqn: RBM Joint Probability}, | |||
<math display="block"> | |||
\begin{equation}\label{eqn: RBM visible probability} | |||
P_{\textrm{rbm}}(\bm{v}) = \sum_{\bm{h}} P_{\textrm{rbm}}(\bm{v},\bm{h}) = \frac{1}{Z}\sum_{h}e^{-E(\bm{v},\bm{h})}. | |||
\end{equation} | |||
</math> | |||
As a result of the restriction, the visible units, with the hidden units fixed, are mutually independent: given a choice of the hidden units <math>\bm{h}</math>, we have an '''independent''' probability distribution for '''each''' visible unit given by | |||
<math display="block"> | |||
\begin{equation} | |||
P_{\textrm{rbm}}(v_{i} = 1 | \bm{h}) = \sigma(a_{i} + \sum_{j}W_{ij}h_{j}), | |||
\qquad i=1,\ldots, n_{\mathrm{v}}, | |||
\end{equation} | |||
</math> | |||
where <math>\sigma(x) = 1/(1+e^{-x})</math> is the sigmoid function. Similarly, with the visible units fixed, the individual hidden units are also mutually independent with the probability distribution | |||
<math display="block"> | |||
\begin{equation}\label{eqn: RBM P(h|v)} | |||
P_{\textrm{rbm}}(h_{j} = 1 | \bm{v}) = \sigma(b_{j} + \sum_{i}v_{i}W_{ij}) | |||
\qquad j=1,\ldots, n_{\mathrm{h}}. | |||
\end{equation} | |||
</math> | |||
The visible (hidden) units can thus be interpreted as artificial neurons connected to the hidden (visible) units with sigmoid activation function and bias <math>\bm{a}</math> (<math>\bm{b}</math>). | |||
A direct consequence of this mutual independence is that sampling a vector <math>\bm{v}</math> or <math>\bm{h}</math> reduces to sampling every component individually. | |||
Notice that this simplification comes about due to the restriction that visible (hidden) units do not directly interact amongst themselves, i.e. there are no terms proportional to <math>v_i v_j</math> or <math>h_i h_j</math> in Eq.~\eqref{eqn: RBM Energy}. | |||
In the following, we explain how one can train an RBM and discuss possible applications of RBMs. | |||
====Training an RBM==== | |||
Consider a set of binary input data <math>\bm{x}_k</math>, <math>k=1,\ldots,M</math>, drawn from a probability distribution <math>P_{\textrm{data}}(\bm{x})</math>. The aim of the training is to tune the parameters <math>\lbrace \bm{a}, \bm{b}, W \rbrace</math> in an RBM such that after training <math>P_{\textrm{rbm}}(\bm{x}) \approx P_{\textrm{data}}(\bm{x})</math>. | |||
The standard approach to solve this problem is the maximum likelihood principle, in other words we want to find the parameters <math>\lbrace \bm{a}, \bm{b}, W \rbrace</math> which maximize the probability that our model produces the data <math>\bm{x}_k</math>. | |||
Maximizing the likelihood <math>\mathcal{L}(\bm{a},\bm{b},W) = \prod P_{\textrm{rbm}}(\bm{x}_{k})</math> is equivalent to training the RBM using a loss function we have encountered before, the negative log-likelihood | |||
<math display="block"> | |||
\begin{equation} | |||
L(\bm{a},\bm{b},W) = - \sum_{k=1}^{M} \log P_{\textrm{rbm}}(\bm{x}_{k}). | |||
\end{equation} | |||
</math> | |||
For the gradient descent, we need derivatives of the loss function | |||
of the form | |||
<math display="block"> | |||
\begin{equation}\label{eqn: log-likelihood derivative} | |||
\frac{\partial L(\bm{a},\bm{b},W)}{\partial W_{ij}} = -\sum_{k=1}^{M} \frac{\partial\log P_{\textrm{rbm}}(\bm{x}_{k})}{\partial W_{ij}}. | |||
\end{equation} | |||
</math> | |||
This derivative consists of two terms, | |||
<math display="block"> | |||
\begin{equation} \label{eqn: RBM derivatives} | |||
\begin{split} | |||
\frac{\partial\log P_{\textrm{rbm}}(\bm{x})}{\partial W_{ij}} &= x_{i}P_{\textrm{rbm}}(h_{j}=1|\bm{x}) - \sum_{\bm{v}} v_{i} P_{\textrm{rbm}}(h_{j}=1|\bm{v}) P_{\textrm{rbm}}(\bm{v}) | |||
\end{split} | |||
\end{equation} | |||
</math> | |||
and similarly simple forms are found for the derivatives with respect to the components of <math>\bm{a}</math> and <math>\bm{b}</math>. | |||
We can then iteratively update the parameters just as we have done in Chapter~[[guide:4b07d6b69a#sec:supervised |sec:supervised]] | |||
, | |||
<math display="block"> | |||
\begin{equation} | |||
W_{ij} \rightarrow W_{ij} - \eta \frac{\partial L(a,b,W)}{\partial W_{ij}} | |||
\end{equation} | |||
</math> | |||
with a sufficiently small learning rate <math>\eta</math>. | |||
As we have seen in the previous chapter in the context of backpropagation, we can reduce the computational cost by replacing the summation over the whole data set in Eq.~\eqref{eqn: log-likelihood derivative} with a summation over a small randomly chosen batch of samples. This reduction in the computational cost comes at the expense of noise, but at the same time it can help to improve generalization. | |||
However, there is one more problem: The second summation in Eq.~\eqref{eqn: RBM derivatives}, which contains <math>2^{n_v}</math> terms, cannot be efficiently evaluated exactly. Instead, we have to approximate the sum by sampling the visible layer <math>\bm{v}</math> from the marginal probability distribution <math>P_{\textrm{rbm}}(\bm{v})</math>. This sampling can be done using ''Gibbs sampling'' as follows: | |||
<proc id="alg: Gibbs Sampling" label = "Gibbs sampling"> | |||
[H] | |||
\SetAlgoLined | |||
<table><tr><td>'''Input:''' </td><td>Any visible vector <math>\bm{v}(0)</math></td></tr></table> | |||
<table><tr><td>'''Output:''' </td><td>Visible vector <math>\bm{v}(r)</math></td></tr></table> | |||
'''for''' <math>n=1</math>\dots <math>r</math> '''do'''<div class="ms-2 border-start border-dark ps-2"> | |||
<div> sample <math>\bm{h}(n)</math> from <math>P_{\rm rbm}(\bm{h}|\bm{v}=\bm{v}(n-1))</math>;</div><div> sample <math>\bm{v}(n)</math> from <math>P_{\rm rbm}(\bm{v}|\bm{h}=\bm{h}(n))</math>;</div></div>'''end''' | |||
</proc> | |||
With sufficiently many steps <math>r</math>, the vector <math>\bm{v}(r)</math> is an unbiased sample drawn from <math>P_{\textrm{rbm}}(\bm{v})</math>. By repeating the procedure, we can obtain multiple samples to estimate the summation. Note that this is still rather computationally expensive, requiring multiple evaluations on the model. | |||
The key innovation which allows the training of an RBM to be computationally feasible was proposed by Geoffrey Hinton (2002). Instead of obtaining multiple samples, we simply perform the Gibbs sampling with <math>r</math> steps and estimate the summation with a single sample, in other words we replace the second summation in Eq.~\eqref{eqn: RBM derivatives} with | |||
<math display="block"> | |||
\begin{equation} | |||
\sum_{\bm{v}} v_{i} P_{\textrm{rbm}}(h_{j}=1|\bm{v}) P_{\textrm{rbm}}(\bm{v}) \rightarrow v'_{i} P_{\textrm{rbm}}(h_{j}=1|\bm{v}'), | |||
\end{equation} | |||
</math> | |||
where <math>\bm{v}' = \bm{v}(r)</math> is simply the sample obtained from <math>r</math>-step Gibbs sampling. With this modification, the gradient, Eq.~\eqref{eqn: RBM derivatives}, can be approximated as | |||
<math display="block"> | |||
\begin{equation} | |||
\frac{\partial\log P_{\textrm{rbm}}(\bm{x})}{\partial W_{ij}} \approx x_{i}P_{\textrm{rbm}}(h_{j}=1|\bm{x}) - v'_{i} P_{\textrm{rbm}}(h_{j}=1|\bm{v}'). | |||
\end{equation} | |||
</math> | |||
This method is known as ''contrastive divergence''. Although the quantity computed is only a biased estimator of the gradient, this approach is found to work well in practice. | |||
The complete algorithm for training a RBM with <math>r</math>-step contrastive divergence can be summarised as follows: | |||
\<proc id="alg: contrastive divergence" label = "Contrastive divergence">[H] | |||
\SetAlgoLined | |||
<table><tr><td>'''Input:''' </td><td>Dataset <math>\mathcal{D} = \lbrace \bm{x}_{1}, \bm{x}_{2}, \dots \bm{x}_{M} \rbrace</math> drawn from a distribution <math>P(x)</math></td></tr></table> | |||
<div>initialize the RBM weights <math>\lbrace \bm{a},\bm{b},W \rbrace</math>;</div><div>Initialize <math>\Delta W_{ij} = \Delta a_{i} = \Delta b_{j} =0</math>;</div>'''while''' not converged '''do'''<div class="ms-2 border-start border-dark ps-2"> | |||
<div> select a random batch <math>S</math> of samples from the dataset <math>\mathcal{D}</math> ;</div> '''forall''' <math>\bm{x} \in S</math> '''do'''<div class="ms-2 border-start border-dark ps-2"> | |||
Obtain <math>\bm{v}'</math> by <math>r</math>-step Gibbs sampling starting from <math>\bm{x}</math> <math>\Delta W_{ij} \leftarrow \Delta W_{ij} - x_{i}P_{\textrm{rbm}}(h_{j}=1|\bm{x}) + v'_{i} P_{\textrm{rbm}}(h_{j}=1|\bm{v}')</math> | |||
</div>'''end''' | |||
<math>W_{ij} \leftarrow W_{ij} - \eta\Delta W_{ij}</math> | |||
(and similarly for <math>\bm{a}</math> and <math>\bm{b}</math>) | |||
</div>'''end''' | |||
</proc> | |||
Having trained the RBM to represent the underlying data distribution <math>P(\bm{x})</math>, there are a few ways one can use the trained model: | |||
<ul><li> '''Pretraining''' We can use <math>W</math> and <math>\bm{b}</math> as the initial weights and biases for a deep network (c.f. Chapter 4), which is then fine-tuned with gradient descent and backpropagation. | |||
</li> | |||
<li> '''Generative Modelling''' As a generative model, a trained RBM can be used to generate new samples via Gibbs sampling (Alg.~[[#alg: Gibbs Sampling |alg: Gibbs Sampling]]). Some potential uses of the generative aspect of the RBM include ''recommender systems'' and ''image reconstruction''. In the following subsection, we provide an example, where an RBM is used to reconstruct a noisy signal. | |||
</li> | |||
</ul> | |||
====Example: signal or image reconstruction/denoising==== | |||
A major drawback of the simple RBMs for their application is the fact that they only take binary data as input. As an example, we thus look at simple periodic waveforms with 60 sample points. In particular, we use sawtooth, sine, and square waveforms. In order to have quasi-continuous data, we use eight bits for each point, such that our signal can take values from 0 to 255. Finally, we generate samples to train with a small variation in the maximum value, the periodicity, as well as the center point of each waveform. | |||
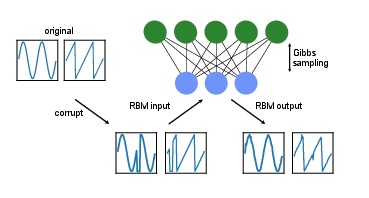
After training the RBM using the contrastive divergence algorithm, we now have a model which represents the data distribution of the binarized waveforms. Consider now a signal which has been corrupted, meaning some parts of the waveform have not been received, in other words they are set to 0. By feeding this corrupted data into the RBM and performing a few iterations of Gibbs sampling (Alg.~[[#alg: Gibbs Sampling |alg: Gibbs Sampling]]), we can obtain a reconstruction of the signal, where the missing part has been repaired, as can been seen at the bottom of Fig.~[[#fig:RBM_reconstruction |fig:RBM_reconstruction]]. | |||
Note that the same procedure can be used to reconstruct or denoise images. Due to the limitation to binary data, however, the picture has to either be binarized, or the input size to the RBM becomes fairly large for high-resolution pictures. It is thus not surprising that while RBMs have been popular in the mid-2000s, they have largely been superseded by more modern and architectures such as ''generative adversarial networks'' which we shall explore later in the chapter. However, they still serve a pedagogical purpose and could also provide inspiration for future innovations, in particular in science. A recent example is the idea of using an RBM to represent a quantum mechanical state. | |||
<div id="fig:RBM_reconstruction" class="d-flex justify-content-center"> | |||
[[File:guide_7132c_rbm_reconstr.png | 400px | thumb | ''' Signal reconstruction.''' Using an RBM to repair a corrupted signal, here a sine and a sawtooth waveform. ]] | |||
</div> | |||
===Training an RNN without supervision=== | |||
<div id="fig:RNN_gen" class="d-flex justify-content-center"> | |||
[[File:guide_7132c_generative_RNN2.png | 400px | thumb | ''' RNN used as a generator.''' For training, left, the input data shifted by one, <math>\bm{x}_{t+1}</math>, are used as the label. For the generation of new sequences, right, we input a single data point <math>\bm{x}_1</math> and the RNN uses the recurrent steps to generate a new sequence. ]] | |||
</div> | |||
In Sec.~[[guide:4b07d6b69a#sec:supervised |sec:supervised]], | |||
the RNN was introduced as a classification model. Instead of classifying sequences of data, such as time series, the RNN can also be trained to generate valid sequences itself. Given the RNN introduced in Sec.~\ref{sec:rnn}, | |||
the implementation of such a generator is straight-forward and does not require a new architecture. The main difference is that the output <math>\bm{y}_t</math> of the network given the data point <math>\bm{x}_t</math> is a guess of the subsequent data point <math>\bm{x}_{t+1}</math> instead of the class to which the whole sequence belongs to. This means in particular that the input and output size are now the same. | |||
For training this network, we can once again use the cross-entropy or (negative) log-likelihood as a loss function, | |||
<math display="block"> | |||
\begin{equation} | |||
L_{\mathrm{ent}} | |||
=-\sum_{t=1}^{m-1} \bm{x}_{t+1}\cdot | |||
\ln \left( | |||
\bm{y}_{t} | |||
\right), | |||
\label{eq:unsup_RNN} | |||
\end{equation} | |||
</math> | |||
where <math>\bm{x}_{t+1}</math> is now the ‘label’ for the input <math>\bm{x}_{t}</math> and <math>\bm{y}_{t}</math> is the output of the network and <math>t</math> runs over the input sequence with length <math>m</math>. This training is schematically shown in Fig.~[[#fig:RNN_gen |fig:RNN_gen]]. | |||
For generating a new sequence, it is enough to have one single input point <math>\bm{x}_1</math> to start the sequence. Note that since we now can start with a single data point <math>\bm{x}_1</math> and generate a whole sequence of data points <math>\{\bm{y}_t\}</math>, this mode of using an RNN is referred to as ''one-to-many''. This sequence generation is shown in Fig.~[[#fig:RNN_gen |fig:RNN_gen]], left. | |||
====<span id="sec:rnn_gen"></span>Example: generating molecules with an RNN==== | |||
To illustrate the concept of sequence generation using recurrent neural networks, we use an RNN to generate new molecules. | |||
The first question we need to address is how to encode a chemical structure into input data---of sequential form no less---that a machine learning model can read. A common representation of molecular graphs used in chemistry is the ''simplified molecular-input line-entry system'', or SMILES. | |||
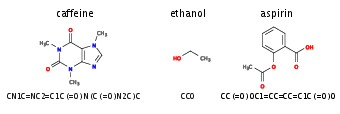
Figure~[[#fig:smiles |fig:smiles]] shows examples of such SMILES strings for the caffeine, ethanol, and aspirin molecules. We can use the dataset ''Molecular Sets'' <ref group="Notes" >[https://github.com/molecularsets/moses]</ref>, which contains <math>\sim 1.9</math>M molecules written in the SMILES format. | |||
Using the SMILES dataset, we create a dictionary to translate each character that appears in the dataset into an integer. We further use one-hot-encoding to feed each character separately to the RNN. This creates a map from characters in SMILES strings onto an array of numbers. | |||
Finally, in order to account for the variable size of the molecules and hence, the variable length of the strings, we can introduce a ‘stop’ character such that the network learns and later generates sequences of arbitrary length. | |||
We are now ready to use the SMILES strings for training our network as described above, where the input is a one-hot-encoded vector and the output is again a vector of the same size. Note, however, that similar to a classification task, the output vector is a probability distribution over the characters the network believes could come next. Unlike a classification task, where we consider the largest output the best guess of the network, here we sample in each step from the probability distribution <math>\bm{y}_t</math> to again have a one-hot-encoded vector for the input of the next step. | |||
<div id="fig:smiles" class="d-flex justify-content-center"> | |||
[[File:guide_7132c_SMILES_examples.png | 400px | thumb | ''' SMILES.''' Examples of molecules and their representation in SMILES. ]] | |||
</div> | |||
===Autoencoders=== | |||
Autoencoders are neuron-based generative models, initially introduced for dimensionality reduction. The original purpose, thus, is similar to that of PCA or t-SNE that we already encountered in Sec.~[[guide:A315bbc881#sec:structuring_data |sec:structuring_data]], | |||
namely the reduction of the number of features that describe our input data. Unlike for PCA, where we have a clear recipe how to reduce the number of features, an autoencoder learns the best way of achieving the dimensionality reduction. An obvious question, however, is how to measure the quality of the compression, which is essential for the definition of a loss function and thus, training. | |||
In the case of t-SNE, we introduced two probability distributions based on the distance of samples in the original and feature space, respectively, and minimized their difference, for example using the Kullback-Leibler divergence. | |||
The solution the autoencoder uses is to have a neural network do first, the dimensionality reduction, or encoding to the ''latent space'', <math>\bm{x}\mapsto \bm{e}(\bm{x})=\bm{z}</math>, and then, the decoding back to the original dimension, <math>\bm{z} \mapsto \bm{d}(\bm{z})</math>, see Fig.~[[#fig:AE_scheme |fig:AE_scheme]]. This architecture allows us to directly compare the original input <math>\bm{x}</math> with the reconstructed output <math>\bm{d}(\bm{e}(\bm{x}))</math>, such that the autoencoder trains itself unsupervised by minimizing the difference. A good example of a loss function that achieves successful training and that we have encountered already several times is the cross entropy, | |||
<math display="block"> | |||
\begin{equation} | |||
L_{\rm ae} = - \sum_i \bm{x}_i \cdot \ln[ \bm{d}(\bm{e}(\bm{x_i}))]. | |||
\end{equation} | |||
</math> | |||
In other words, we compare point-wise the difference between the input to the encoder with the decoder's output. | |||
Intuitively, the latent space with its lower dimension presents a bottleneck for the information propagation from input to output. The goal of training is to find and keep the most relevant information for the reconstruction to be optimal. The latent space then corresponds to the reduced space in PCA and t-SNE. Note that much like in t-SNE but unlike in PCA, the new features are in general not independent. | |||
<div id="fig:AE_scheme" class="d-flex justify-content-center"> | |||
[[File:guide_7132c_autoencoder.png | 400px | thumb | ''' General autoencoder architecture.''' A neural network is used to contract a compressed representation of the input in the latent space. A second neural network is used to reconstruct the original input. ]] | |||
</div> | |||
====Variational autoencoders==== | |||
A major problem of the approach introduced in the previous section is its tendency to overfitting. As an extreme example, a sufficiently complicated encoder-decoder pair could learn to map all data in the training set onto a single variable and back to the data. Such a network would indeed accomplish completely lossless compression and decompression. However, the network would not have extracted any useful information from the dataset and thus, would completely fail to compress and decompress previously unseen data. Moreover, as in the case of the dimensionality-reduction schemes discussed in Sec.~[[guide:A315bbc881#sec:structuring_data |sec:structuring_data]], we would like to analyze the latent space images and extract new information about the data. Finally, we might also want to use the decoder part of the autoencoder as a generator for new data. For these reasons, it is essential that we combat overfitting as we have done in the previous chapters by regularization. | |||
The question then becomes how one can effectively regularize the autoencoder. First, we need to analyze what properties we would like the latent space to fulfil. We can identify two main properties: | |||
<ul><li> If two input data points are close (according to some measure), their images in the latent space should also be close. We call this property ''continuity''. | |||
</li> | |||
<li> Any point in the latent space should be mapped through the decoder onto a meaningful data point, a property we call ''completeness''. | |||
</li> | |||
</ul> | |||
While there are principle ways to achieve regularization along similar paths as discussed in the previous section on supervised learning, we will discuss here a solution that is particularly useful as a generative model: the ''variational autoencoder'' (VAE). | |||
<div id="fig:VAE" class="d-flex justify-content-center"> | |||
[[File:guide_7132c_vae.png | 400px | thumb | ''' Architecture of variational autoencoder.''' Instead of outputting a point <math>z</math> in the latent space, the encoder provides a distribution <math>N(\bm{\mu}, \bm{\sigma})</math>, parametrized by the means <math>\bm{\mu}</math> and the standard deviations <math>\bm{\sigma}</math>. The input <math>\bm{z}</math> for the decoder is then drawn from <math>N(\bm{\mu}, \bm{\sigma})</math>. ]] | |||
</div> | |||
The idea behind VAEs is for the encoder to output not just an exact point <math>\bm{z}</math> in the latent space, but a (factorized) Normal distribution of points, <math>\mathcal{N}(\bm{\mu}, \bm{\sigma})</math>. In particular, the output of the encoder comprises two vectors, the first representing the means, <math>\bm{\mu}</math>, and the second the standard deviations, <math>\bm{\sigma}</math>. The input for the decoder is then sampled from this distribution, <math>\bm{z} \sim \mathcal{N}(\bm{\mu}, \bm{\sigma})</math>, and the original input is reconstructed and compared to the original input for training. In addition to the standard loss function comparing input and output of the VAE, we further add a regularization term to the loss function such that the distributions from the encoder are close to a standard normal distribution <math>\mathcal{N}(\bm{0}, \bm{1})</math>. Using the Kullback-Leibler divergence, Eq.(2.20), | |||
to measure the deviation from the standard normal distribution, the full loss function then reads | |||
<math display="block"> | |||
\begin{align} | |||
L_{\rm vae} &= -\sum_i \bm{x}^{\rm in}_i \ln \bm{x}^{\rm out}_i + {\rm KL} (\mathcal{N}(\bm{\mu}_i, \bm{\sigma}_i)|| \mathcal{N}(\bm{0}, \bm{1}))\nonumber | |||
&= -\sum_i \bm{x}^{\rm in}_i \ln \bm{x}^{\rm out}_i + \frac12 \sum_k [\sigma_{i,k}^2 + \mu_{i,k}^2 -1 -2 \ln\sigma_{i,k}]. | |||
\label{eq:loss_vae} | |||
\end{align} | |||
</math> | |||
In this expression, the first term quantifies the reconstruction loss with <math>\bm{x}_i^{\rm in}</math> the input to and <math>\bm{x}_i^{\rm out}</math> the reconstructed data from the VAE. The second term is the regularization on the latent space for each input data point, which for two (diagonal) Normal distributions can be simplified, see second line of Eq.~\eqref{eq:loss_vae}. | |||
This procedure regularizes the training through the introduction of noise, similar to the dropout layer in Section~[[guide:4b07d6b69a#sec:supervised |sec:supervised]]. | |||
However, the regularization here not only generically increases generalization, but also enforces the desired structure in the latent space. | |||
The structure of a VAE is shown in Fig.~[[#fig:VAE |fig:VAE]]. By enforcing the mean and variance structure of the encoder output, the latent space fulfills the requirements outlined above. This type of structure can then serve as a generative model for many different data types: anything from human faces to complicated molecular structures. Hence, the variational autoencoder goes beyond extracting information from a dataset, but can be used for the scientific discovery. Note, finally, that the general structure of the variational autoencoder can be applied beyond the simple example above. As an example, a different distribution function can be enforced in the latent space other than the standard Normal distribution, or a different neural network can be used as encoder and decoder, such as a RNN. | |||
===Generative adversarial networks=== | |||
In this section, we will be a concerned with a type of generative neural network, the generative adversarial network (GAN), which gained a very high popularity in recent years. | |||
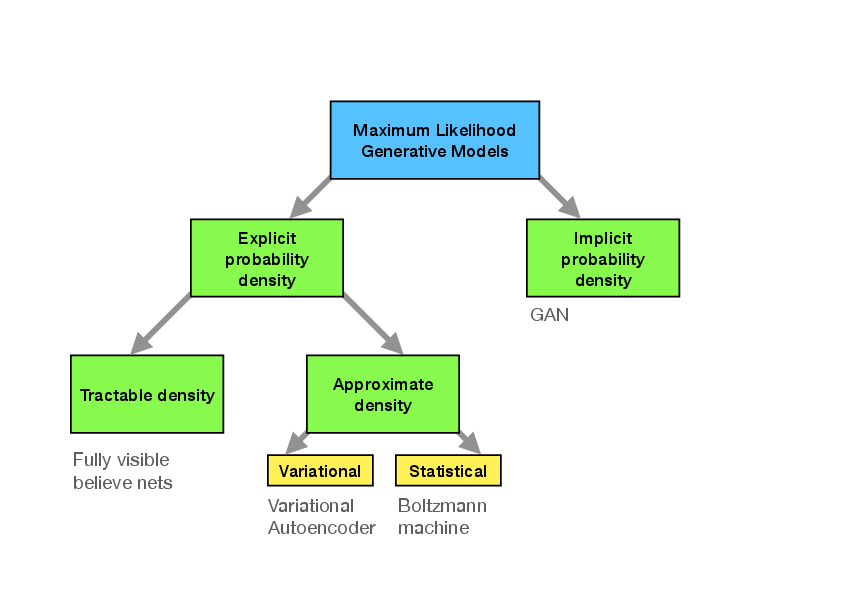
Before getting into the details about this method, we give a quick systematic overview over types of generative methods, to place GANs in proper relation to them.~<ref group="Notes" >following “NIPS 2016 Tutorial: Generative Adversarial Netoworks” Ian Goodfellow, arXiv:1701.001160</ref>. | |||
====Types of generative models==== | |||
<div id="fig: generative taxonomy" class="d-flex justify-content-center"> | |||
[[File:guide_7132c_GenerativeTaxonomy.png | 400px | thumb | ''' Maximum likelihood approaches to generative modeling.''' ]] | |||
</div> | |||
We restrict ourselves to methods that are based on the ''maximum likelihood principle''. The role of the model is to provide an estimate <math>p_{\text{model}}(\bm{x};\bm{\theta})</math> of a probability distribution parametrized by parameters <math>\bm{\theta}</math>. The likelihood is the probability that the model assigns to the training data | |||
<math display="block"> | |||
\begin{equation} | |||
\prod_{i=1}^mp_{\text{model}}(\bm{x}_{i};\bm{\theta}), | |||
\end{equation} | |||
</math> | |||
where <math>m</math> is again the number of samples in the data <math>\{\bm{x}_i\}</math>. The goal is to choose the parameters <math>\bm{\theta}</math> such as to maximize the likelihood. Thanks to the equality | |||
<math display="block"> | |||
\begin{equation} | |||
\begin{split} | |||
\bm{\theta}^*=&\,\underset{\bm{\theta}}{\text{argmax}}\prod_{i=1}^mp_{\text{model}}(\bm{x}_{i};\bm{\theta}) | |||
=&\,\underset{\bm{\theta}}{\text{argmax}}\sum_{i=1}^m\mathrm{log}\,p_{\text{model}}(\bm{x}_{i};\bm{\theta}) | |||
\end{split} | |||
\end{equation} | |||
</math> | |||
we can just as well work with the sum of logarithms, which is easier to handle. As we explained previously (see section on t-SNE), the maximization is equivalent to the minimization of the cross-entropy between two probability distributions: the ‘true’ distribution <math>p_{\mathrm{data}}(\bm{x})</math> from which the data has been drawn and <math>p_{\text{model}}(\bm{x};\bm{\theta})</math>. While we do not have access to <math>p_{\mathrm{data}}(\bm{x})</math> in principle, we estimate it empirically as a distribution peaked at the <math>m</math> data points we have. | |||
Methods can now be distinguished by the way <math>p_{\mathrm{model}}</math> is defined and evaluated (see Fig.~[[#fig: generative taxonomy |fig: generative taxonomy]]). We differentiate between models that define <math>p_{\mathrm{data}}(\bm{x})</math> ''explicitly'' through some functional form. They have the general advantage that maximization of the likelihood is rather straight-forward, since we have direct access to this function. The downside is that the functional forms are generically limiting the ability of the model to fit the data distribution or become computationally intractable. | |||
Among those explicit density models, we can further distinguish between those that represent a computationally tractable density and those that do not. An example for tractable explicit density models are ''fully visible belief networks'' (FVBNs) that decompose the probability distribution over an <math>n</math>-dimensional vector <math>\bm{x}</math> into a product of conditional probabilities | |||
<math display="block"> | |||
\begin{equation} | |||
p_{\mathrm{model}}(\bm{x})=\prod_{j=1}^n\, p_{\mathrm{model}}(x_j|x_1,\cdots,x_{j-1}). | |||
\end{equation} | |||
</math> | |||
We can already see that, once we use the model to draw new samples, this is done one entry of the vector <math>\bm{x}</math> at a time (first <math>x_1</math> is drawn, then, knowing it, <math>x_2</math> is drawn etc.). This is computationally costly and not parallelizable but is useful for tasks that are anyway sequential (like generation of human speech, where the so-called WaveNet employs FVBNs). | |||
Models that encode an explicit density, but require approximations to maximize the likelihood that can either be variational in nature or use stochastic methods. We have seen examples for either. Variational methods define a lower bound to the log likelihood which can be maximized | |||
<math display="block"> | |||
\begin{equation} | |||
\mathcal{L}(\bm{x};\bm{\theta})\leq \mathrm{log}\,p_{\text{model}}(\bm{x};\bm{\theta}). | |||
\end{equation} | |||
</math> | |||
The algorithm produces a maximum value of the log-likelihood that is at least as high as the value for <math>\mathcal{L}</math> obtained (when summed over all data points). Variational autoencoders belong to this category. Their most obvious shortcoming is that <math>\mathcal{L}(\bm{x};\bm{\theta})</math> may represent a very bad lower bound to the log-likelihood (and is in general not guaranteed to converge to it for infinite model size), so that the distribution represented by the model is very different from <math>p_{\mathrm{data}}</math>. | |||
Stochastic methods, in contrast, often rely on a Markov chain process: The model is defined by a probability <math>q(\bm{x}'|\bm{x})</math> from which the current sample <math>\bm{x}'</math> is drawn, which depends on the previously drawn sample <math>\bm{x}</math> (but not any others). RBMs are an example for this. They have the advantage that there is some rigorously proven convergence to <math>p_{\text{model}}</math> with large enough size of the RBM, but the convergence may be slow. Like with FVBNs, the drawing process is sequential and thus not easily parallelizable. | |||
All these classes of models allow for explicit representations of the probability density function approximations. | |||
In contrast, for GANs and related models, there is only an indirect access to said probability density: The model allows us to sample from it. Naturally, this makes optimization potentially harder, but circumvents many of the other previously mentioned problems. In particular | |||
<ul style{{=}}"list-style-type:lower-roman"><li>GANs can generate samples in parallel</li> | |||
<li>there are few restrictions on the form of the generator function (as compared to Boltzmann machines, for instance, which have a restricted form to make Markov chain sampling work)</li> | |||
<li>no Markov chains are needed</li> | |||
<li>no variational bound is needed and some GAN model families are known to be asymptotically consistent (meaning that for a large enough model they are approximations to any probability distribution).</li> | |||
</ul> | |||
GANs have been immensely successful in several application scenarios. Their superiority against other methods is, however, often assessed subjectively. Most of performance comparison have been in the field of image generation, and largely on the ImageNet database. Some of the standard tasks evaluated in this context are: | |||
<ul style{{=}}"list-style-type:lower-roman"><li>generate an image from a sentence or phrase that describes its content (“a blue flower”)</li> | |||
<li>generate realistic images from sketches</li> | |||
<li>generate abstract maps from satellite photos</li> | |||
<li>generate a high-resolution (“super-resolution”) image from a lower resolution one</li> | |||
<li>predict a next frame in a video.</li> | |||
</ul> | |||
As far as more science-related applications are concerned, GANs have been used to | |||
<ul style{{=}}"list-style-type:lower-roman"><li>predict the impact of climate change on individual houses</li> | |||
<li>generate new molecules that have been later synethsized.</li> | |||
</ul> | |||
In the light of these examples, it is of fundamental importance to understand that GANs enable (and excel at) problems with multi-modal outputs. That means the problems are such that a single input corresponds to many different ‘correct’ or ‘likely’ outputs. (In contrast to a mathematical function, which would always produce the same output.) This is important to keep in mind in particular in scientific applications, where we often search for ''the one answer''. Only if that is not the case, GANs can actually play out their strengths. | |||
Let us consider image super-resolution as an illustrative example: Conventional (deterministic) methods of increasing image resolution would necessarily lead to some blurring or artifacts, because the information that can be encoded in the finer pixel grid simply is not existent in the input data. A GAN, in contrast, will provide a possibility how a realistic image could have looked if it had been taken with higher resolution. This way they add information that may differ from the true scene of the image that was taken -- a process that is obviously not yielding a unique answer since many versions of the information added may correspond to a realistic image. | |||
<div id="fig:GAN_scheme" class="d-flex justify-content-center"> | |||
[[File:guide_7132c_GAN.png | 400px | thumb | ''' Architecture of a GAN.''' ]] | |||
</div> | |||
====The working principle of GANs==== | |||
The optimization of all neural network models we discussed so far was formulated as minimization of a cost function. For GANs, while such a formulation is a also possible, a much more illuminating perspective is viewing the GAN as a ''game'' between two players, the ''generator'' (<math>G</math>) and the ''discriminator'' (<math>D</math>), see Fig.~[[#fig:GAN_scheme |fig:GAN_scheme]]. The role of <math>G</math> is to generate from some random input <math>\bm{z}</math> drawn from a simple distribution samples that could be mistaken from being drawn from <math>p_{\mathrm{data}}</math>. The task of <math>D</math> is to classify its input as generated by <math>G</math> or coming from the data. Training should improve the performance of both <math>D</math> and <math>G</math> at their respective tasks simultaneously. After training is completed, <math>G</math> can be used to draw samples that closely resembles those drawn from <math>p_{\mathrm{data}}</math>. | |||
In summary | |||
<math display="block"> | |||
\begin{equation} | |||
\begin{split} | |||
D_{\bm{\theta}_D}&:\ \bm{x}\mapsto \text{binary true/false}, | |||
G_{\bm{\theta}_G}&:\ \bm{z}\mapsto \bm{x}, | |||
\end{split} | |||
\end{equation} | |||
</math> | |||
where we have also indicated the two sets of parameters on which the two functions depend: <math>\bm{\theta}_D</math> and <math>\bm{\theta}_G</math>, respectively. | |||
The game is then defined by two cost functions. The discriminator wants to minimize <math>J_D(\bm{\theta}_D,\bm{\theta}_G)</math> by only changing <math>\bm{\theta}_D</math>, while the generator <math>J_G(\bm{\theta}_D,\bm{\theta}_G)</math> by only changing <math>\bm{\theta}_G</math>. So, each players cost depends on both their and the other players parameters, the latter of which cannot be controlled by the player. The solution to this game optimization problem is a (local) minimum, i.e., a point in <math>(\bm{\theta}_D,\bm{\theta}_G)</math>-space where <math>J_D(\bm{\theta}_D,\bm{\theta}_G)</math> has a local minimum with respect to <math>\bm{\theta}_D</math> and <math>J_G(\bm{\theta}_D,\bm{\theta}_G)</math> has a local minimum with respect to <math>\bm{\theta}_G</math>. In game theory such a solution is called a Nash equilibrium. | |||
Let us now specify possible choices for the cost functions as well as for <math>D</math> and <math>G</math>. | |||
The most important requirement of <math>G</math> is that it is differentiable. It thus can (in contrast to VAEs) not have discrete variables on the output layer. A typical representation is a deep (possibly convolutional) neural network. A popular Deep Conventional architecture is called DCGAN. Then <math>\bm{\theta}_G</math> are the networks weights and biases. The input <math>\bm{z}</math> is drawn from some simple prior distribution, e.g., the uniform distribution or a normal distribution. (The specific choice of this distribution is secondary, as long as we use the same during training and when we use the generator by itself.) It is important that <math>\bm{z}</math> has at least as high a dimension as <math>\bm{x}</math> if the full multi-dimensional <math>p_{\text{model}}</math> is to be approximated. Otherwise the model will perform some sort of dimensional reduction. Several tweaks have also been used, such as feeding some components of <math>\bm{z}</math> into a hidden instead of the input layer and adding noise to hidden layers. | |||
The training proceeds in steps. At each step, a minibatch of <math>\bm{x}</math> is drawn from the data set and a minibatch of <math>\bm{z}</math> is sampled from the prior distribution. Using this, gradient descent-type updates are performed: One update of <math>\bm{\theta}_D</math> using the gradient of <math>J_D(\bm{\theta}_D,\bm{\theta}_G)</math> and one of <math>\bm{\theta}_G</math> using the gradient of <math>J_G(\bm{\theta}_D,\bm{\theta}_G)</math>. | |||
====The cost functions==== | |||
For <math>D</math>, the cost function of choice is the cross-entropy as with standard binary classifiers that have sigmoid output. Given that the labels are ‘1’ for data and ‘0’ for <math>\bm{z}</math> samples, it is simply | |||
<math display="block"> | |||
\begin{equation} | |||
J_D(\bm{\theta}_D,\bm{\theta}_G) | |||
=-\frac{1}{2 N_1}\sum_i\,\log\,D(\bm{x}_i)-\frac{1}{2 N_2}\sum_j\log (1-D(G(\bm{z}_j))), | |||
\end{equation} | |||
</math> | |||
where the sums over <math>i</math> and <math>j</math> run over the respective minibatches, which contain <math>N_1</math> and <math>N_2</math> points. | |||
For <math>G</math> more variations of the cost functions have been explored. Maybe the most intuitive one is | |||
<math display="block"> | |||
\begin{equation} | |||
J_G(\bm{\theta}_D,\bm{\theta}_G)=-J_D(\bm{\theta}_D,\bm{\theta}_G), | |||
\end{equation} | |||
</math> | |||
which corresponds to the so-called ''zero-sum'' or ''minmax'' game. Its solution is formally given by | |||
<math display="block"> | |||
\begin{equation} | |||
\bm{\theta}_G^\star=\underset{\bm{\theta}_G}{\text{arg min}}\ \ \underset{\bm{\theta}_D}{\text{max}} | |||
\left[-J_D(\bm{\theta}_D,\bm{\theta}_G)\right]. | |||
\label{eq: GAN Minmax} | |||
\end{equation} | |||
</math> | |||
This form of the cost is convenient for theoretical analysis, because there is only a single target function, which helps drawing parallels to conventional optimization. However, other cost functions have been proven superior in practice. The reason is that minimization can get trapped very far from an equilibrium: When the discriminator manages to learn rejecting generator samples with high confidence, the gradient of the generator will be very small, making its optimization very hard. | |||
Instead, we can use the cross-entropy also for the generator cost function (but this time from the generator's perspective) | |||
<math display="block"> | |||
\begin{equation} | |||
J_G(\bm{\theta}_D,\bm{\theta}_G)=-\frac{1}{2 N_2}\sum_j\log\, D(G(\bm{z}_j)). | |||
\end{equation} | |||
</math> | |||
Now the generator maximizes the probability of the discriminator being mistaken. This way, each player still has a strong gradient when the player is loosing the game. We observe that this version of <math>J_G(\bm{\theta}_D,\bm{\theta}_G)</math> has no direct dependence of the training data. Of course, such a dependence is implicit via <math>D</math>, which has learned from the training data. This indirect dependence also acts like a regularizer, preventing overfitting: <math>G</math> has no possibility to directly ‘fit’ its output to training data. | |||
====Remarks==== | |||
In closing this section, we comment on a few properties of GANs, which also mark frontiers for improvements. One global problem is that GANs are typically difficult to train: they require large training sets and are highly susceptible to hyper-parameter fluctuations. It is currently an active topic of research to compensate for this with the structural modification and novel loss function formulations. | |||
'''Mode collapse ---''' this may describe one of the most obvious problems of GANs: it refers to a situation where <math>G</math> does not explore the full space to which <math>\bm{x}</math> belongs, but rather maps several inputs <math>\bm{z}</math> to the same output. Mode collapse can be more or less severe. For instance a <math>G</math> trained on generating images may always resort to certain fragments or patterns of images. A formal reason for mode collapse is when the simultaneous gradient descent gravitates towards a solution | |||
<math display="block"> | |||
\begin{equation} | |||
\bm{\theta}_G^\star=\underset{\bm{\theta}_D}{\text{arg max}}\ \ \underset{\bm{\theta}_G}{\text{min}} | |||
\left[-J_D(\bm{\theta}_D,\bm{\theta}_G)\right], | |||
\end{equation} | |||
</math> | |||
instead of the order in Eq.~\eqref{eq: GAN Minmax}. (A priori it is not clear which of the two solutions is closer to the algorithm's doing.) Note that the interchange of min and max in general corresponds to a different solution: It is now sufficient for <math>G</math> to always produce one (and the same) output that is classified as data by <math>D</math> with very high probability. Due to the mode collapse problem, GANs are not good at exploring ergodically the full space of possible outputs. They rather produce few very good possible outputs. | |||
One strategy to fight mode collapse is called ''minibatch features''. Instead of letting <math>D</math> rate one sample at a time, a minibatch of real and generated samples is considered at once. It then detects whether the generated samples are unusually close to each other. | |||
'''Arithmetics with GANs ---''' it has been demonstrated that GANs can do linear arithmetics with inputs to add or remove abstract features from the output. This has been demonstrated using a DCGAN trained on images of faces. The gender and the feature ‘wearing glasses’ can be added or subtracted and thus changed at will. Of course such a result is only empirical, there is no formal mathematical theory why it works. | |||
'''Using GANs with labelled data ---''' it has been shown that, if (partially) labeled data is available, using the labels when training <math>D</math> may improve the performance of <math>G</math>. In this constellation, <math>G</math> has still the same task as before and does not interact with the labels. If data with <math>n</math> classes exist, then <math>D</math> will be constructed as a classifier for <math>(n+1)</math> classes, where the extra class corresponds to ‘fake’ data that <math>D</math> attributes to coming from <math>G</math>. If a data point has a label, then this label is used as a reference in the cost function. If a datapoint has no label, then the first <math>n</math> outputs of <math>D</math> are summed up. | |||
'''One-sided label smoothing ---''' | |||
this technique is useful not only for the <math>D</math> in GANs but also other binary classification problems with neural networks. Often we observe that classifiers give proper results, but show a too confident probability. This overshooting confidence can be counteracted by one-sided label smoothing. The idea is to simply replace the target value for the real examples with a value slightly less than 1, e.g., 0.9. This smoothes the distribution of the discriminator. Why do we only perform this off-set one-sided and not also give a small nonzero value <math>\beta</math> to the fake samples target values? If we were to do this, the optimal function for <math>D</math> is | |||
<math display="block"> | |||
\begin{equation} | |||
D^\star(\bm{x})=\frac{p_{\mathrm{data}}(\bm{x})+\beta p_{\mathrm{model}}(\bm{x})}{p_{\mathrm{data}}(\bm{x})+ p_{\mathrm{model}}(\bm{x})}. | |||
\end{equation} | |||
</math> | |||
Consider now a range of <math>\bm{x}</math> for which <math>p_{\mathrm{data}}(\bm{x})</math> is small but <math>p_{\mathrm{model}}(\bm{x})</math> is large (a “spurious mode”). <math>D^\star(\bm{x})</math> will have a peak near this spurious mode. This means <math>D</math> reinforces incorrect behavior of <math>G</math>. This will encourage <math>G</math> to reproduce samples that it already makes (irrespective of whether they are anything like real data). | |||
==General references== | |||
{{cite arXiv|last1=Neupert|first1=Titus|last2=Fischer|first2=Mark H|last3=Greplova|first3=Eliska|last4=Choo|first4=Kenny|last5=Denner|first5=M. Michael|year=2022|title=Introduction to Machine Learning for the Sciences|eprint=2102.04883|class=physics.comp-ph}} | |||
==Notes== | |||
{{Reflist|group=Notes}} | |||
Revision as of 01:32, 17 May 2024
\label{sec:unsupervised} In Sec.~sec:supervised, we discussed supervised learning tasks, for which datasets consist of input-output pairs, or data-label pairs. More often than not, however, we have data without labels and would like to extract information from such a dataset. Clustering problems fall in this category, for instance: We suspect that the data can be divided into different types, but we do not know which features distinguish these types. Mathematically, we can think of the data [math]\bm{x}[/math] as samples that were drawn from a probability distribution [math]P(\bm{x})[/math]. The unsupervised learning task is to implicitly represent this distribution with a model, for example represented by a neural network. The model can then be used to study properties of the distribution or to generate new ‘artificial’ data. The models we encounter in this chapter are thus also referred to as generative models. In general, unsupervised learning is conceptually more challenging than supervised learning. At the same time, unsupervised algorithms are highly desirable, since unlabelled data is much more abundant than labelled data. Moreover, we can in principle use a generative model for a classification task by learning the joint probability distribution of the data-label pair. In this chapter, we will introduce three types of neural networks that are specific to unsupervised learning tasks: Restricted Boltzmann machines, autoencoders, and generative adversarial networks. Furthermore, we will discuss how the RNN introduced in the previous chapter can also be used for an unsupervised task.
Restricted Boltzmann machine
Restricted Boltzmann Machines (RBM) are a class of generative stochastic neural networks. More specifically, given some (binary) input data [math]\bm{x}\in\{0,1\}^{n_v}[/math], an RBM can be trained to approximate the probability distribution of this input. Moreover, once the neural network is trained to approximate the distribution of the input, we can sample from the network, in other words we generate new instances from the learned probability distribution. The RBM consists of two layers (see Fig.~fig:RBM) of binary units. Each binary unit is a variable which can take the values [math]0[/math] or [math]1[/math]. We call the first (input) layer visible and the second layer hidden. The visible layer with input variables [math]\lbrace v_{1}, v_{2}, \dots v_{n_{\mathrm{v}}}\rbrace [/math], which we collect in the vector [math]\bm{v}[/math], is connected to the hidden layer with variables [math]\{ h_{1}, h_{2}, \dots h_{n_{\mathrm{h}}}\}[/math], which we collect in the vector [math]\bm{h}[/math]. The role of the hidden layer is to mediate correlations between the units of the visible layer. In contrast to the neural networks we have seen in the previous chapter, the hidden layer is not followed by an output layer. Instead, the RBM represents a probability distribution [math]P_{\text{rbm}}(\bm{v})[/math], which depends on variational parameters represented by the weights and biases of a neural network. The RBM, as illustrated by the graph in Fig.~fig:RBM, is a special case of a network structure known as a Boltzmann machine with the restriction that a unit in the visible layer is only connected to hidden units and vice versa, hence the name restricted Boltzmann machine.
The structure of the RBM is motivated from statistical physics: To each choice of the binary vectors [math]\bm{v}[/math] and [math]\bm{h}[/math], we assign a value we call the energy
where the vectors [math]\bm{a}[/math], [math]\bm{b}[/math], and the matrix [math]W[/math] are the variational parameters of the model. Given the energy, the probability distribution over the configurations [math](\bm{v}, \bm{h})[/math] is defined as
where
is a normalisation factor called the partition function. The sum in Eq.~\eqref{eq: partition function} runs over all binary vectors [math]\bm{v}[/math] and [math]\bm{h}[/math], i.e., vectors with entries [math]0[/math] or [math]1[/math]. The probability that the model assigns to a visible vector [math]\bm{v}[/math] is then the marginal over the joint probability distribution Eq.~\eqref{eqn: RBM Joint Probability},
As a result of the restriction, the visible units, with the hidden units fixed, are mutually independent: given a choice of the hidden units [math]\bm{h}[/math], we have an independent probability distribution for each visible unit given by
where [math]\sigma(x) = 1/(1+e^{-x})[/math] is the sigmoid function. Similarly, with the visible units fixed, the individual hidden units are also mutually independent with the probability distribution
The visible (hidden) units can thus be interpreted as artificial neurons connected to the hidden (visible) units with sigmoid activation function and bias [math]\bm{a}[/math] ([math]\bm{b}[/math]). A direct consequence of this mutual independence is that sampling a vector [math]\bm{v}[/math] or [math]\bm{h}[/math] reduces to sampling every component individually. Notice that this simplification comes about due to the restriction that visible (hidden) units do not directly interact amongst themselves, i.e. there are no terms proportional to [math]v_i v_j[/math] or [math]h_i h_j[/math] in Eq.~\eqref{eqn: RBM Energy}. In the following, we explain how one can train an RBM and discuss possible applications of RBMs.
Training an RBM
Consider a set of binary input data [math]\bm{x}_k[/math], [math]k=1,\ldots,M[/math], drawn from a probability distribution [math]P_{\textrm{data}}(\bm{x})[/math]. The aim of the training is to tune the parameters [math]\lbrace \bm{a}, \bm{b}, W \rbrace[/math] in an RBM such that after training [math]P_{\textrm{rbm}}(\bm{x}) \approx P_{\textrm{data}}(\bm{x})[/math]. The standard approach to solve this problem is the maximum likelihood principle, in other words we want to find the parameters [math]\lbrace \bm{a}, \bm{b}, W \rbrace[/math] which maximize the probability that our model produces the data [math]\bm{x}_k[/math]. Maximizing the likelihood [math]\mathcal{L}(\bm{a},\bm{b},W) = \prod P_{\textrm{rbm}}(\bm{x}_{k})[/math] is equivalent to training the RBM using a loss function we have encountered before, the negative log-likelihood
For the gradient descent, we need derivatives of the loss function of the form
This derivative consists of two terms,
and similarly simple forms are found for the derivatives with respect to the components of [math]\bm{a}[/math] and [math]\bm{b}[/math]. We can then iteratively update the parameters just as we have done in Chapter~sec:supervised ,
with a sufficiently small learning rate [math]\eta[/math]. As we have seen in the previous chapter in the context of backpropagation, we can reduce the computational cost by replacing the summation over the whole data set in Eq.~\eqref{eqn: log-likelihood derivative} with a summation over a small randomly chosen batch of samples. This reduction in the computational cost comes at the expense of noise, but at the same time it can help to improve generalization. However, there is one more problem: The second summation in Eq.~\eqref{eqn: RBM derivatives}, which contains [math]2^{n_v}[/math] terms, cannot be efficiently evaluated exactly. Instead, we have to approximate the sum by sampling the visible layer [math]\bm{v}[/math] from the marginal probability distribution [math]P_{\textrm{rbm}}(\bm{v})[/math]. This sampling can be done using Gibbs sampling as follows:
[H] \SetAlgoLined
| Input: | Any visible vector [math]\bm{v}(0)[/math] |
| Output: | Visible vector [math]\bm{v}(r)[/math] |
With sufficiently many steps [math]r[/math], the vector [math]\bm{v}(r)[/math] is an unbiased sample drawn from [math]P_{\textrm{rbm}}(\bm{v})[/math]. By repeating the procedure, we can obtain multiple samples to estimate the summation. Note that this is still rather computationally expensive, requiring multiple evaluations on the model. The key innovation which allows the training of an RBM to be computationally feasible was proposed by Geoffrey Hinton (2002). Instead of obtaining multiple samples, we simply perform the Gibbs sampling with [math]r[/math] steps and estimate the summation with a single sample, in other words we replace the second summation in Eq.~\eqref{eqn: RBM derivatives} with
where [math]\bm{v}' = \bm{v}(r)[/math] is simply the sample obtained from [math]r[/math]-step Gibbs sampling. With this modification, the gradient, Eq.~\eqref{eqn: RBM derivatives}, can be approximated as
This method is known as contrastive divergence. Although the quantity computed is only a biased estimator of the gradient, this approach is found to work well in practice.
The complete algorithm for training a RBM with [math]r[/math]-step contrastive divergence can be summarised as follows:
\
\SetAlgoLined
| Input: | Dataset [math]\mathcal{D} = \lbrace \bm{x}_{1}, \bm{x}_{2}, \dots \bm{x}_{M} \rbrace[/math] drawn from a distribution [math]P(x)[/math] |
Obtain [math]\bm{v}'[/math] by [math]r[/math]-step Gibbs sampling starting from [math]\bm{x}[/math] [math]\Delta W_{ij} \leftarrow \Delta W_{ij} - x_{i}P_{\textrm{rbm}}(h_{j}=1|\bm{x}) + v'_{i} P_{\textrm{rbm}}(h_{j}=1|\bm{v}')[/math]
[math]W_{ij} \leftarrow W_{ij} - \eta\Delta W_{ij}[/math] (and similarly for [math]\bm{a}[/math] and [math]\bm{b}[/math])
Having trained the RBM to represent the underlying data distribution [math]P(\bm{x})[/math], there are a few ways one can use the trained model:
- Pretraining We can use [math]W[/math] and [math]\bm{b}[/math] as the initial weights and biases for a deep network (c.f. Chapter 4), which is then fine-tuned with gradient descent and backpropagation.
- Generative Modelling As a generative model, a trained RBM can be used to generate new samples via Gibbs sampling (Alg.~alg: Gibbs Sampling). Some potential uses of the generative aspect of the RBM include recommender systems and image reconstruction. In the following subsection, we provide an example, where an RBM is used to reconstruct a noisy signal.
Example: signal or image reconstruction/denoising
A major drawback of the simple RBMs for their application is the fact that they only take binary data as input. As an example, we thus look at simple periodic waveforms with 60 sample points. In particular, we use sawtooth, sine, and square waveforms. In order to have quasi-continuous data, we use eight bits for each point, such that our signal can take values from 0 to 255. Finally, we generate samples to train with a small variation in the maximum value, the periodicity, as well as the center point of each waveform. After training the RBM using the contrastive divergence algorithm, we now have a model which represents the data distribution of the binarized waveforms. Consider now a signal which has been corrupted, meaning some parts of the waveform have not been received, in other words they are set to 0. By feeding this corrupted data into the RBM and performing a few iterations of Gibbs sampling (Alg.~alg: Gibbs Sampling), we can obtain a reconstruction of the signal, where the missing part has been repaired, as can been seen at the bottom of Fig.~fig:RBM_reconstruction. Note that the same procedure can be used to reconstruct or denoise images. Due to the limitation to binary data, however, the picture has to either be binarized, or the input size to the RBM becomes fairly large for high-resolution pictures. It is thus not surprising that while RBMs have been popular in the mid-2000s, they have largely been superseded by more modern and architectures such as generative adversarial networks which we shall explore later in the chapter. However, they still serve a pedagogical purpose and could also provide inspiration for future innovations, in particular in science. A recent example is the idea of using an RBM to represent a quantum mechanical state.
Training an RNN without supervision
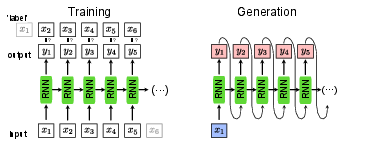
In Sec.~sec:supervised, the RNN was introduced as a classification model. Instead of classifying sequences of data, such as time series, the RNN can also be trained to generate valid sequences itself. Given the RNN introduced in Sec.~\ref{sec:rnn}, the implementation of such a generator is straight-forward and does not require a new architecture. The main difference is that the output [math]\bm{y}_t[/math] of the network given the data point [math]\bm{x}_t[/math] is a guess of the subsequent data point [math]\bm{x}_{t+1}[/math] instead of the class to which the whole sequence belongs to. This means in particular that the input and output size are now the same. For training this network, we can once again use the cross-entropy or (negative) log-likelihood as a loss function,
where [math]\bm{x}_{t+1}[/math] is now the ‘label’ for the input [math]\bm{x}_{t}[/math] and [math]\bm{y}_{t}[/math] is the output of the network and [math]t[/math] runs over the input sequence with length [math]m[/math]. This training is schematically shown in Fig.~fig:RNN_gen. For generating a new sequence, it is enough to have one single input point [math]\bm{x}_1[/math] to start the sequence. Note that since we now can start with a single data point [math]\bm{x}_1[/math] and generate a whole sequence of data points [math]\{\bm{y}_t\}[/math], this mode of using an RNN is referred to as one-to-many. This sequence generation is shown in Fig.~fig:RNN_gen, left.
Example: generating molecules with an RNN
To illustrate the concept of sequence generation using recurrent neural networks, we use an RNN to generate new molecules. The first question we need to address is how to encode a chemical structure into input data---of sequential form no less---that a machine learning model can read. A common representation of molecular graphs used in chemistry is the simplified molecular-input line-entry system, or SMILES. Figure~fig:smiles shows examples of such SMILES strings for the caffeine, ethanol, and aspirin molecules. We can use the dataset Molecular Sets [Notes 1], which contains [math]\sim 1.9[/math]M molecules written in the SMILES format. Using the SMILES dataset, we create a dictionary to translate each character that appears in the dataset into an integer. We further use one-hot-encoding to feed each character separately to the RNN. This creates a map from characters in SMILES strings onto an array of numbers. Finally, in order to account for the variable size of the molecules and hence, the variable length of the strings, we can introduce a ‘stop’ character such that the network learns and later generates sequences of arbitrary length. We are now ready to use the SMILES strings for training our network as described above, where the input is a one-hot-encoded vector and the output is again a vector of the same size. Note, however, that similar to a classification task, the output vector is a probability distribution over the characters the network believes could come next. Unlike a classification task, where we consider the largest output the best guess of the network, here we sample in each step from the probability distribution [math]\bm{y}_t[/math] to again have a one-hot-encoded vector for the input of the next step.
Autoencoders
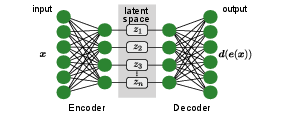
Autoencoders are neuron-based generative models, initially introduced for dimensionality reduction. The original purpose, thus, is similar to that of PCA or t-SNE that we already encountered in Sec.~sec:structuring_data, namely the reduction of the number of features that describe our input data. Unlike for PCA, where we have a clear recipe how to reduce the number of features, an autoencoder learns the best way of achieving the dimensionality reduction. An obvious question, however, is how to measure the quality of the compression, which is essential for the definition of a loss function and thus, training. In the case of t-SNE, we introduced two probability distributions based on the distance of samples in the original and feature space, respectively, and minimized their difference, for example using the Kullback-Leibler divergence. The solution the autoencoder uses is to have a neural network do first, the dimensionality reduction, or encoding to the latent space, [math]\bm{x}\mapsto \bm{e}(\bm{x})=\bm{z}[/math], and then, the decoding back to the original dimension, [math]\bm{z} \mapsto \bm{d}(\bm{z})[/math], see Fig.~fig:AE_scheme. This architecture allows us to directly compare the original input [math]\bm{x}[/math] with the reconstructed output [math]\bm{d}(\bm{e}(\bm{x}))[/math], such that the autoencoder trains itself unsupervised by minimizing the difference. A good example of a loss function that achieves successful training and that we have encountered already several times is the cross entropy,
In other words, we compare point-wise the difference between the input to the encoder with the decoder's output. Intuitively, the latent space with its lower dimension presents a bottleneck for the information propagation from input to output. The goal of training is to find and keep the most relevant information for the reconstruction to be optimal. The latent space then corresponds to the reduced space in PCA and t-SNE. Note that much like in t-SNE but unlike in PCA, the new features are in general not independent.
Variational autoencoders
A major problem of the approach introduced in the previous section is its tendency to overfitting. As an extreme example, a sufficiently complicated encoder-decoder pair could learn to map all data in the training set onto a single variable and back to the data. Such a network would indeed accomplish completely lossless compression and decompression. However, the network would not have extracted any useful information from the dataset and thus, would completely fail to compress and decompress previously unseen data. Moreover, as in the case of the dimensionality-reduction schemes discussed in Sec.~sec:structuring_data, we would like to analyze the latent space images and extract new information about the data. Finally, we might also want to use the decoder part of the autoencoder as a generator for new data. For these reasons, it is essential that we combat overfitting as we have done in the previous chapters by regularization. The question then becomes how one can effectively regularize the autoencoder. First, we need to analyze what properties we would like the latent space to fulfil. We can identify two main properties:
- If two input data points are close (according to some measure), their images in the latent space should also be close. We call this property continuity.
- Any point in the latent space should be mapped through the decoder onto a meaningful data point, a property we call completeness.
While there are principle ways to achieve regularization along similar paths as discussed in the previous section on supervised learning, we will discuss here a solution that is particularly useful as a generative model: the variational autoencoder (VAE).
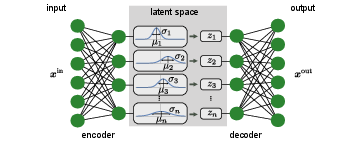
The idea behind VAEs is for the encoder to output not just an exact point [math]\bm{z}[/math] in the latent space, but a (factorized) Normal distribution of points, [math]\mathcal{N}(\bm{\mu}, \bm{\sigma})[/math]. In particular, the output of the encoder comprises two vectors, the first representing the means, [math]\bm{\mu}[/math], and the second the standard deviations, [math]\bm{\sigma}[/math]. The input for the decoder is then sampled from this distribution, [math]\bm{z} \sim \mathcal{N}(\bm{\mu}, \bm{\sigma})[/math], and the original input is reconstructed and compared to the original input for training. In addition to the standard loss function comparing input and output of the VAE, we further add a regularization term to the loss function such that the distributions from the encoder are close to a standard normal distribution [math]\mathcal{N}(\bm{0}, \bm{1})[/math]. Using the Kullback-Leibler divergence, Eq.(2.20), to measure the deviation from the standard normal distribution, the full loss function then reads
In this expression, the first term quantifies the reconstruction loss with [math]\bm{x}_i^{\rm in}[/math] the input to and [math]\bm{x}_i^{\rm out}[/math] the reconstructed data from the VAE. The second term is the regularization on the latent space for each input data point, which for two (diagonal) Normal distributions can be simplified, see second line of Eq.~\eqref{eq:loss_vae}. This procedure regularizes the training through the introduction of noise, similar to the dropout layer in Section~sec:supervised. However, the regularization here not only generically increases generalization, but also enforces the desired structure in the latent space.
The structure of a VAE is shown in Fig.~fig:VAE. By enforcing the mean and variance structure of the encoder output, the latent space fulfills the requirements outlined above. This type of structure can then serve as a generative model for many different data types: anything from human faces to complicated molecular structures. Hence, the variational autoencoder goes beyond extracting information from a dataset, but can be used for the scientific discovery. Note, finally, that the general structure of the variational autoencoder can be applied beyond the simple example above. As an example, a different distribution function can be enforced in the latent space other than the standard Normal distribution, or a different neural network can be used as encoder and decoder, such as a RNN.
Generative adversarial networks
In this section, we will be a concerned with a type of generative neural network, the generative adversarial network (GAN), which gained a very high popularity in recent years. Before getting into the details about this method, we give a quick systematic overview over types of generative methods, to place GANs in proper relation to them.~[Notes 2].
Types of generative models
We restrict ourselves to methods that are based on the maximum likelihood principle. The role of the model is to provide an estimate [math]p_{\text{model}}(\bm{x};\bm{\theta})[/math] of a probability distribution parametrized by parameters [math]\bm{\theta}[/math]. The likelihood is the probability that the model assigns to the training data
where [math]m[/math] is again the number of samples in the data [math]\{\bm{x}_i\}[/math]. The goal is to choose the parameters [math]\bm{\theta}[/math] such as to maximize the likelihood. Thanks to the equality
we can just as well work with the sum of logarithms, which is easier to handle. As we explained previously (see section on t-SNE), the maximization is equivalent to the minimization of the cross-entropy between two probability distributions: the ‘true’ distribution [math]p_{\mathrm{data}}(\bm{x})[/math] from which the data has been drawn and [math]p_{\text{model}}(\bm{x};\bm{\theta})[/math]. While we do not have access to [math]p_{\mathrm{data}}(\bm{x})[/math] in principle, we estimate it empirically as a distribution peaked at the [math]m[/math] data points we have. Methods can now be distinguished by the way [math]p_{\mathrm{model}}[/math] is defined and evaluated (see Fig.~fig: generative taxonomy). We differentiate between models that define [math]p_{\mathrm{data}}(\bm{x})[/math] explicitly through some functional form. They have the general advantage that maximization of the likelihood is rather straight-forward, since we have direct access to this function. The downside is that the functional forms are generically limiting the ability of the model to fit the data distribution or become computationally intractable. Among those explicit density models, we can further distinguish between those that represent a computationally tractable density and those that do not. An example for tractable explicit density models are fully visible belief networks (FVBNs) that decompose the probability distribution over an [math]n[/math]-dimensional vector [math]\bm{x}[/math] into a product of conditional probabilities
We can already see that, once we use the model to draw new samples, this is done one entry of the vector [math]\bm{x}[/math] at a time (first [math]x_1[/math] is drawn, then, knowing it, [math]x_2[/math] is drawn etc.). This is computationally costly and not parallelizable but is useful for tasks that are anyway sequential (like generation of human speech, where the so-called WaveNet employs FVBNs). Models that encode an explicit density, but require approximations to maximize the likelihood that can either be variational in nature or use stochastic methods. We have seen examples for either. Variational methods define a lower bound to the log likelihood which can be maximized
The algorithm produces a maximum value of the log-likelihood that is at least as high as the value for [math]\mathcal{L}[/math] obtained (when summed over all data points). Variational autoencoders belong to this category. Their most obvious shortcoming is that [math]\mathcal{L}(\bm{x};\bm{\theta})[/math] may represent a very bad lower bound to the log-likelihood (and is in general not guaranteed to converge to it for infinite model size), so that the distribution represented by the model is very different from [math]p_{\mathrm{data}}[/math]. Stochastic methods, in contrast, often rely on a Markov chain process: The model is defined by a probability [math]q(\bm{x}'|\bm{x})[/math] from which the current sample [math]\bm{x}'[/math] is drawn, which depends on the previously drawn sample [math]\bm{x}[/math] (but not any others). RBMs are an example for this. They have the advantage that there is some rigorously proven convergence to [math]p_{\text{model}}[/math] with large enough size of the RBM, but the convergence may be slow. Like with FVBNs, the drawing process is sequential and thus not easily parallelizable.
All these classes of models allow for explicit representations of the probability density function approximations. In contrast, for GANs and related models, there is only an indirect access to said probability density: The model allows us to sample from it. Naturally, this makes optimization potentially harder, but circumvents many of the other previously mentioned problems. In particular
- GANs can generate samples in parallel
- there are few restrictions on the form of the generator function (as compared to Boltzmann machines, for instance, which have a restricted form to make Markov chain sampling work)
- no Markov chains are needed
- no variational bound is needed and some GAN model families are known to be asymptotically consistent (meaning that for a large enough model they are approximations to any probability distribution).
GANs have been immensely successful in several application scenarios. Their superiority against other methods is, however, often assessed subjectively. Most of performance comparison have been in the field of image generation, and largely on the ImageNet database. Some of the standard tasks evaluated in this context are:
- generate an image from a sentence or phrase that describes its content (“a blue flower”)
- generate realistic images from sketches
- generate abstract maps from satellite photos
- generate a high-resolution (“super-resolution”) image from a lower resolution one
- predict a next frame in a video.
As far as more science-related applications are concerned, GANs have been used to
- predict the impact of climate change on individual houses
- generate new molecules that have been later synethsized.
In the light of these examples, it is of fundamental importance to understand that GANs enable (and excel at) problems with multi-modal outputs. That means the problems are such that a single input corresponds to many different ‘correct’ or ‘likely’ outputs. (In contrast to a mathematical function, which would always produce the same output.) This is important to keep in mind in particular in scientific applications, where we often search for the one answer. Only if that is not the case, GANs can actually play out their strengths. Let us consider image super-resolution as an illustrative example: Conventional (deterministic) methods of increasing image resolution would necessarily lead to some blurring or artifacts, because the information that can be encoded in the finer pixel grid simply is not existent in the input data. A GAN, in contrast, will provide a possibility how a realistic image could have looked if it had been taken with higher resolution. This way they add information that may differ from the true scene of the image that was taken -- a process that is obviously not yielding a unique answer since many versions of the information added may correspond to a realistic image.
The working principle of GANs
The optimization of all neural network models we discussed so far was formulated as minimization of a cost function. For GANs, while such a formulation is a also possible, a much more illuminating perspective is viewing the GAN as a game between two players, the generator ([math]G[/math]) and the discriminator ([math]D[/math]), see Fig.~fig:GAN_scheme. The role of [math]G[/math] is to generate from some random input [math]\bm{z}[/math] drawn from a simple distribution samples that could be mistaken from being drawn from [math]p_{\mathrm{data}}[/math]. The task of [math]D[/math] is to classify its input as generated by [math]G[/math] or coming from the data. Training should improve the performance of both [math]D[/math] and [math]G[/math] at their respective tasks simultaneously. After training is completed, [math]G[/math] can be used to draw samples that closely resembles those drawn from [math]p_{\mathrm{data}}[/math]. In summary
where we have also indicated the two sets of parameters on which the two functions depend: [math]\bm{\theta}_D[/math] and [math]\bm{\theta}_G[/math], respectively. The game is then defined by two cost functions. The discriminator wants to minimize [math]J_D(\bm{\theta}_D,\bm{\theta}_G)[/math] by only changing [math]\bm{\theta}_D[/math], while the generator [math]J_G(\bm{\theta}_D,\bm{\theta}_G)[/math] by only changing [math]\bm{\theta}_G[/math]. So, each players cost depends on both their and the other players parameters, the latter of which cannot be controlled by the player. The solution to this game optimization problem is a (local) minimum, i.e., a point in [math](\bm{\theta}_D,\bm{\theta}_G)[/math]-space where [math]J_D(\bm{\theta}_D,\bm{\theta}_G)[/math] has a local minimum with respect to [math]\bm{\theta}_D[/math] and [math]J_G(\bm{\theta}_D,\bm{\theta}_G)[/math] has a local minimum with respect to [math]\bm{\theta}_G[/math]. In game theory such a solution is called a Nash equilibrium. Let us now specify possible choices for the cost functions as well as for [math]D[/math] and [math]G[/math]. The most important requirement of [math]G[/math] is that it is differentiable. It thus can (in contrast to VAEs) not have discrete variables on the output layer. A typical representation is a deep (possibly convolutional) neural network. A popular Deep Conventional architecture is called DCGAN. Then [math]\bm{\theta}_G[/math] are the networks weights and biases. The input [math]\bm{z}[/math] is drawn from some simple prior distribution, e.g., the uniform distribution or a normal distribution. (The specific choice of this distribution is secondary, as long as we use the same during training and when we use the generator by itself.) It is important that [math]\bm{z}[/math] has at least as high a dimension as [math]\bm{x}[/math] if the full multi-dimensional [math]p_{\text{model}}[/math] is to be approximated. Otherwise the model will perform some sort of dimensional reduction. Several tweaks have also been used, such as feeding some components of [math]\bm{z}[/math] into a hidden instead of the input layer and adding noise to hidden layers. The training proceeds in steps. At each step, a minibatch of [math]\bm{x}[/math] is drawn from the data set and a minibatch of [math]\bm{z}[/math] is sampled from the prior distribution. Using this, gradient descent-type updates are performed: One update of [math]\bm{\theta}_D[/math] using the gradient of [math]J_D(\bm{\theta}_D,\bm{\theta}_G)[/math] and one of [math]\bm{\theta}_G[/math] using the gradient of [math]J_G(\bm{\theta}_D,\bm{\theta}_G)[/math].
The cost functions
For [math]D[/math], the cost function of choice is the cross-entropy as with standard binary classifiers that have sigmoid output. Given that the labels are ‘1’ for data and ‘0’ for [math]\bm{z}[/math] samples, it is simply
where the sums over [math]i[/math] and [math]j[/math] run over the respective minibatches, which contain [math]N_1[/math] and [math]N_2[/math] points. For [math]G[/math] more variations of the cost functions have been explored. Maybe the most intuitive one is
which corresponds to the so-called zero-sum or minmax game. Its solution is formally given by
This form of the cost is convenient for theoretical analysis, because there is only a single target function, which helps drawing parallels to conventional optimization. However, other cost functions have been proven superior in practice. The reason is that minimization can get trapped very far from an equilibrium: When the discriminator manages to learn rejecting generator samples with high confidence, the gradient of the generator will be very small, making its optimization very hard. Instead, we can use the cross-entropy also for the generator cost function (but this time from the generator's perspective)
Now the generator maximizes the probability of the discriminator being mistaken. This way, each player still has a strong gradient when the player is loosing the game. We observe that this version of [math]J_G(\bm{\theta}_D,\bm{\theta}_G)[/math] has no direct dependence of the training data. Of course, such a dependence is implicit via [math]D[/math], which has learned from the training data. This indirect dependence also acts like a regularizer, preventing overfitting: [math]G[/math] has no possibility to directly ‘fit’ its output to training data.
Remarks
In closing this section, we comment on a few properties of GANs, which also mark frontiers for improvements. One global problem is that GANs are typically difficult to train: they require large training sets and are highly susceptible to hyper-parameter fluctuations. It is currently an active topic of research to compensate for this with the structural modification and novel loss function formulations.
Mode collapse --- this may describe one of the most obvious problems of GANs: it refers to a situation where [math]G[/math] does not explore the full space to which [math]\bm{x}[/math] belongs, but rather maps several inputs [math]\bm{z}[/math] to the same output. Mode collapse can be more or less severe. For instance a [math]G[/math] trained on generating images may always resort to certain fragments or patterns of images. A formal reason for mode collapse is when the simultaneous gradient descent gravitates towards a solution
instead of the order in Eq.~\eqref{eq: GAN Minmax}. (A priori it is not clear which of the two solutions is closer to the algorithm's doing.) Note that the interchange of min and max in general corresponds to a different solution: It is now sufficient for [math]G[/math] to always produce one (and the same) output that is classified as data by [math]D[/math] with very high probability. Due to the mode collapse problem, GANs are not good at exploring ergodically the full space of possible outputs. They rather produce few very good possible outputs. One strategy to fight mode collapse is called minibatch features. Instead of letting [math]D[/math] rate one sample at a time, a minibatch of real and generated samples is considered at once. It then detects whether the generated samples are unusually close to each other.
Arithmetics with GANs --- it has been demonstrated that GANs can do linear arithmetics with inputs to add or remove abstract features from the output. This has been demonstrated using a DCGAN trained on images of faces. The gender and the feature ‘wearing glasses’ can be added or subtracted and thus changed at will. Of course such a result is only empirical, there is no formal mathematical theory why it works.
Using GANs with labelled data --- it has been shown that, if (partially) labeled data is available, using the labels when training [math]D[/math] may improve the performance of [math]G[/math]. In this constellation, [math]G[/math] has still the same task as before and does not interact with the labels. If data with [math]n[/math] classes exist, then [math]D[/math] will be constructed as a classifier for [math](n+1)[/math] classes, where the extra class corresponds to ‘fake’ data that [math]D[/math] attributes to coming from [math]G[/math]. If a data point has a label, then this label is used as a reference in the cost function. If a datapoint has no label, then the first [math]n[/math] outputs of [math]D[/math] are summed up.
One-sided label smoothing --- this technique is useful not only for the [math]D[/math] in GANs but also other binary classification problems with neural networks. Often we observe that classifiers give proper results, but show a too confident probability. This overshooting confidence can be counteracted by one-sided label smoothing. The idea is to simply replace the target value for the real examples with a value slightly less than 1, e.g., 0.9. This smoothes the distribution of the discriminator. Why do we only perform this off-set one-sided and not also give a small nonzero value [math]\beta[/math] to the fake samples target values? If we were to do this, the optimal function for [math]D[/math] is
Consider now a range of [math]\bm{x}[/math] for which [math]p_{\mathrm{data}}(\bm{x})[/math] is small but [math]p_{\mathrm{model}}(\bm{x})[/math] is large (a “spurious mode”). [math]D^\star(\bm{x})[/math] will have a peak near this spurious mode. This means [math]D[/math] reinforces incorrect behavior of [math]G[/math]. This will encourage [math]G[/math] to reproduce samples that it already makes (irrespective of whether they are anything like real data).
General references
Neupert, Titus; Fischer, Mark H; Greplova, Eliska; Choo, Kenny; Denner, M. Michael (2022). "Introduction to Machine Learning for the Sciences". arXiv:2102.04883 [physics.comp-ph].
Notes
- [1]
- following “NIPS 2016 Tutorial: Generative Adversarial Netoworks” Ian Goodfellow, arXiv:1701.001160