guide:F3c4e12d38: Difference between revisions
No edit summary |
mNo edit summary |
||
| Line 66: | Line 66: | ||
</math> | </math> | ||
where the sum runs over the <math>m</math> samples of the dataset. This loss function is sometimes also called the ''L2-loss'' and can be seen as a measure of the distance between the output values from the dataset <math>y_i</math> and the corresponding predictions <math>f(\bm{x}_i|\bm{\beta})</math>. | where the sum runs over the <math>m</math> samples of the dataset. This loss function is sometimes also called the ''L2-loss'' and can be seen as a measure of the distance between the output values from the dataset <math>y_i</math> and the corresponding predictions <math>f(\bm{x}_i|\bm{\beta})</math>. | ||
It is convenient to define the <math>m</math> by <math>(n+1)</math> data matrix <math>\widetilde{X}</math>, each row of which corresponds to an input sample <math>\bm{\tilde{x}}^{T}_{i}</math>, as well as the output vector <math>\bm{Y}^{T} = (y_{1}, \dots, y_{m})</math>. With this notation, Eq. | It is convenient to define the <math>m</math> by <math>(n+1)</math> data matrix <math>\widetilde{X}</math>, each row of which corresponds to an input sample <math>\bm{\tilde{x}}^{T}_{i}</math>, as well as the output vector <math>\bm{Y}^{T} = (y_{1}, \dots, y_{m})</math>. With this notation, Eq.\eqref{eqn: RSS} can be expressed succinctly as a matrix equation | ||
<math display="block"> | <math display="block"> | ||
| Line 103: | Line 103: | ||
\end{equation} | \end{equation} | ||
</math> | </math> | ||
where the parameters <math>\beta_{jk}</math> now have an additional index <math>k = 1, \dots, p</math>. Considering the parameters <math>\beta</math> as a <math>(n+1)</math> by <math>p</math> matrix, we can show that the solution takes the same form as before [Eq. | where the parameters <math>\beta_{jk}</math> now have an additional index <math>k = 1, \dots, p</math>. Considering the parameters <math>\beta</math> as a <math>(n+1)</math> by <math>p</math> matrix, we can show that the solution takes the same form as before [Eq.\eqref{eqn: LG RSS Solution}] with <math>Y</math> as a <math>m</math> by <math>p</math> output matrix. | ||
====Statistical analysis==== | ====Statistical analysis==== | ||
| Line 117: | Line 117: | ||
\end{equation} | \end{equation} | ||
</math> | </math> | ||
We assume that this error <math>\epsilon</math> is a Gaussian random variable with mean <math>\mu = 0</math> and variance <math>\sigma^2</math>, which we denote by <math>\epsilon \sim \mathcal{N}(0, \sigma^2)</math>. Assuming that a linear model in Eq. | We assume that this error <math>\epsilon</math> is a Gaussian random variable with mean <math>\mu = 0</math> and variance <math>\sigma^2</math>, which we denote by <math>\epsilon \sim \mathcal{N}(0, \sigma^2)</math>. Assuming that a linear model in Eq.\eqref{eqn: Univariate Linear Model} is a suitable model for our dataset, we are interested in the following question: How does our solution <math>\hat{\bm{\beta}}</math> as given in Eq.\eqref{eqn: LG RSS Solution} compare with the true solution <math>\bm{\beta}^{\textrm{true}}</math> which obeys | ||
<math display="block"> | <math display="block"> | ||
| Line 128: | Line 128: | ||
</math> | </math> | ||
In order to make statistical statements about this question, we have to imagine that we can fix the inputs <math>\bm{x}_{i}</math> of our dataset and repeatedly draw samples for our outputs <math>y_i</math>. Each time we will obtain a different value for <math>y_i</math> following Eq. | In order to make statistical statements about this question, we have to imagine that we can fix the inputs <math>\bm{x}_{i}</math> of our dataset and repeatedly draw samples for our outputs <math>y_i</math>. Each time we will obtain a different value for <math>y_i</math> following Eq.\eqref{eqn: True Linear}, in other words the <math>\epsilon_i</math> are uncorrelated random numbers. | ||
This allows us to formalise the notion of an ''expectation value'' <math>E(\cdots)</math> as the average over an infinite number of draws. | This allows us to formalise the notion of an ''expectation value'' <math>E(\cdots)</math> as the average over an infinite number of draws. | ||
For each draw, we obtain a new dataset, which differs from the other ones by the values of the outputs <math>y_i</math>. With each of these datasets, we obtain a different solution <math>\hat{\bm{\beta}}</math> as given by Eq. | For each draw, we obtain a new dataset, which differs from the other ones by the values of the outputs <math>y_i</math>. With each of these datasets, we obtain a different solution <math>\hat{\bm{\beta}}</math> as given by Eq.\eqref{eqn: LG RSS Solution}. The expectation value <math>E(\hat{\bm{\beta}})</math> is then simply the average value we obtained across an infinite number of datasets. The deviation of this average value from the “true” value given perfect data is called the ''bias'' of the model, | ||
<math display="block"> | <math display="block"> | ||
| Line 149: | Line 149: | ||
\end{equation} | \end{equation} | ||
</math> | </math> | ||
where the second line follows because <math>E(\bm{\epsilon}) = \bm{0}</math> and <math>(\widetilde{X}^{T}\widetilde{X})^{-1} \widetilde{X}^{T} \bm{Y}^{\textrm{true}} = \bm{\beta}^{\textrm{true}}</math>. Equation | where the second line follows because <math>E(\bm{\epsilon}) = \bm{0}</math> and <math>(\widetilde{X}^{T}\widetilde{X})^{-1} \widetilde{X}^{T} \bm{Y}^{\textrm{true}} = \bm{\beta}^{\textrm{true}}</math>. Equation\eqref{eqn: LG RSS Unbiased} implies linear regression is unbiased. Note that other machine learning algorithms will in general be biased. | ||
What about the standard error or uncertainty of our solution? This information is contained in the ''covariance matrix'' | What about the standard error or uncertainty of our solution? This information is contained in the ''covariance matrix'' | ||
| Line 159: | Line 159: | ||
\end{equation} | \end{equation} | ||
</math> | </math> | ||
The covariance matrix can be computed for the case of linear regression using the solution in Eq. | The covariance matrix can be computed for the case of linear regression using the solution in Eq.\eqref{eqn: LG RSS Solution}, the expectation value in Eq.\eqref{eqn: LG RSS Unbiased} and the assumption in Eq.\eqref{eqn: True Linear} that <math>Y = Y^{\textrm{true}} + \bm{\epsilon}</math> yielding | ||
<math display="block"> | <math display="block"> | ||
| Line 193: | Line 193: | ||
</math> | </math> | ||
of the individual parameters <math>\beta_i</math>. The standard error or uncertainty is then <math>\sqrt{\textrm{Var}(\hat{\beta}_{i})}</math>. | of the individual parameters <math>\beta_i</math>. The standard error or uncertainty is then <math>\sqrt{\textrm{Var}(\hat{\beta}_{i})}</math>. | ||
There is one more missing element: we have not explained how to obtain the variances <math>\sigma^2</math> of the outputs <math>y</math>. In an actual machine learning task, we would not know anything about the true relation, as given in Eq. | There is one more missing element: we have not explained how to obtain the variances <math>\sigma^2</math> of the outputs <math>y</math>. In an actual machine learning task, we would not know anything about the true relation, as given in Eq.\eqref{eqn: True Linear}, governing our dataset. The only information we have access to is a single dataset. Therefore, we have to estimate the variance using the samples in our dataset, which is given by | ||
<math display="block"> | <math display="block"> | ||
| Line 222: | Line 222: | ||
\end{equation} | \end{equation} | ||
</math> | </math> | ||
Based on the assumption that the dataset indeed derives from a linear model as given by Eq. | Based on the assumption that the dataset indeed derives from a linear model as given by Eq.\eqref{eqn: True Linear} with a Gaussian error, it can be shown that the RSS solution, Eq.\eqref{eqn: LG RSS Solution}, gives the minimum error among all unbiased linear estimators, Eq.\eqref{eqn: Univariate Linear Model}. | ||
This is known as the Gauss-Markov theorem. | This is known as the Gauss-Markov theorem. | ||
This completes our error analysis of the method. | This completes our error analysis of the method. | ||
====Regularization and the bias-variance tradeoff==== | ====Regularization and the bias-variance tradeoff==== | ||
Although the RSS solution has the minimum error among unbiased linear estimators, the expression for the generalisation error, Eq. | Although the RSS solution has the minimum error among unbiased linear estimators, the expression for the generalisation error, Eq.\eqref{eqn: MSE Generalisation Error}, suggests that we can actually still reduce the error by sacrificing some bias in our estimate. | ||
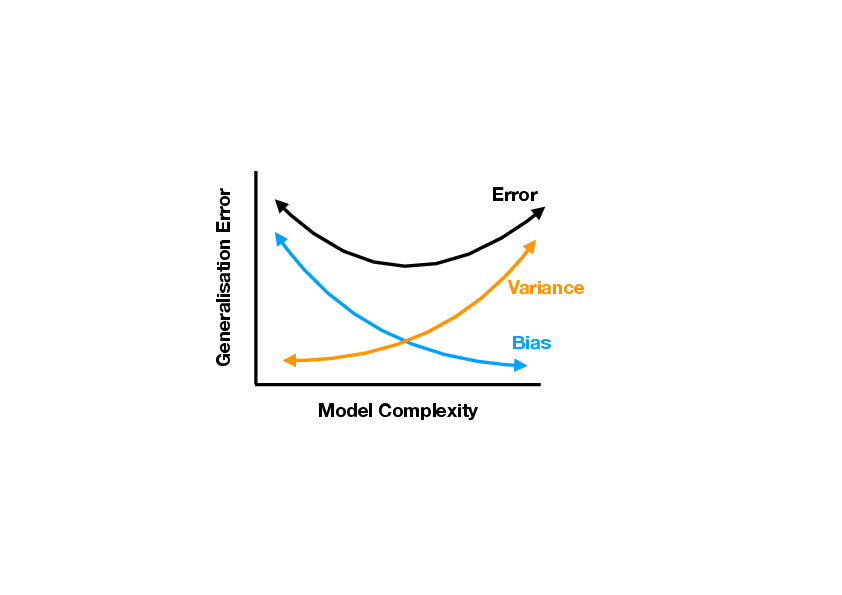
<div id="fig: Bias-Variance Tradeoff" class="d-flex justify-content-center"> | <div id="fig: Bias-Variance Tradeoff" class="d-flex justify-content-center"> | ||
[[File:guide_7132c_Bias-Variance-Tradeoff.png | 400px | thumb | '''Schematic depiction of the bias-variance tradeoff.''' ]] | [[File:guide_7132c_Bias-Variance-Tradeoff.png | 400px | thumb | '''Schematic depiction of the bias-variance tradeoff.''' ]] | ||
| Line 239: | Line 239: | ||
</math> | </math> | ||
This is equivalent to fixing some parameters to zero, i.e., <math>\beta_k = 0</math> if <math>x_{k} \notin \mathcal{M}</math>. Minimizing the RSS with this constraint results in a biased estimator but the reduction in model variance can sometimes help to reduce the overall generalisation error. For a small number of features <math>n \sim 20</math>, one can search exhaustively for the best subset of features that minimises the error, but beyond that the search becomes computationally unfeasible. | This is equivalent to fixing some parameters to zero, i.e., <math>\beta_k = 0</math> if <math>x_{k} \notin \mathcal{M}</math>. Minimizing the RSS with this constraint results in a biased estimator but the reduction in model variance can sometimes help to reduce the overall generalisation error. For a small number of features <math>n \sim 20</math>, one can search exhaustively for the best subset of features that minimises the error, but beyond that the search becomes computationally unfeasible. | ||
A common alternative is called ''ridge regression''. In this method, we consider the same linear model given in Eq. | A common alternative is called ''ridge regression''. In this method, we consider the same linear model given in Eq.\eqref{eqn: Univariate Linear Model} but with a modified loss function | ||
<math display="block"> | <math display="block"> | ||
| Line 247: | Line 247: | ||
</math> | </math> | ||
where <math>\lambda > 0</math> is a positive parameter. | where <math>\lambda > 0</math> is a positive parameter. | ||
This is almost the same as the RSS apart from the term proportional to <math>\lambda</math> [c.f. Eq. | This is almost the same as the RSS apart from the term proportional to <math>\lambda</math> [c.f. Eq.\eqref{eqn: RSS}]. The effect of this new term is to penalize large parameters <math>\beta_j</math> and bias the model towards smaller absolute values. The parameter <math>\lambda</math> is an example of a ''hyper-parameter''\index{hyper-parameter}, which is kept fixed during the training. On fixing <math>\lambda</math> and minimising the loss function, we obtain the solution | ||
<math display="block"> | <math display="block"> | ||
| Line 266: | Line 266: | ||
</math> | </math> | ||
it is also obvious that increasing <math>\lambda</math> increases the bias, while reducing the variance. This is the tradeoff between bias and variance. By appropriately choosing <math>\lambda</math> it is possible that generalisation error can be reduced. We will introduce in the next section a common strategy how to find the optimal value for <math>\lambda</math>. | it is also obvious that increasing <math>\lambda</math> increases the bias, while reducing the variance. This is the tradeoff between bias and variance. By appropriately choosing <math>\lambda</math> it is possible that generalisation error can be reduced. We will introduce in the next section a common strategy how to find the optimal value for <math>\lambda</math>. | ||
The techniques presented here to reduce the generalization error, namely dropping of features and biasing the model to small parameters, are part of a large class of methods known as ''regularization''. Comparing the two methods, we can see a similarity. Both methods actually reduce the complexity of our model. In the former, some parameters are set to zero, while in the latter, there is a constraint which effectively reduces the magnitude of all parameters. A less complex model has a smaller variance but larger bias. By balancing these competing effects, generalisation can be improved, as illustrated schematically in Fig. | The techniques presented here to reduce the generalization error, namely dropping of features and biasing the model to small parameters, are part of a large class of methods known as ''regularization''. Comparing the two methods, we can see a similarity. Both methods actually reduce the complexity of our model. In the former, some parameters are set to zero, while in the latter, there is a constraint which effectively reduces the magnitude of all parameters. A less complex model has a smaller variance but larger bias. By balancing these competing effects, generalisation can be improved, as illustrated schematically in Fig.[[#fig: Bias-Variance Tradeoff |fig: Bias-Variance Tradeoff]]. | ||
In the next chapter, we will see that these techniques are useful beyond applications to linear methods. We illustrate the different concepts in the following example. | In the next chapter, we will see that these techniques are useful beyond applications to linear methods. We illustrate the different concepts in the following example. | ||
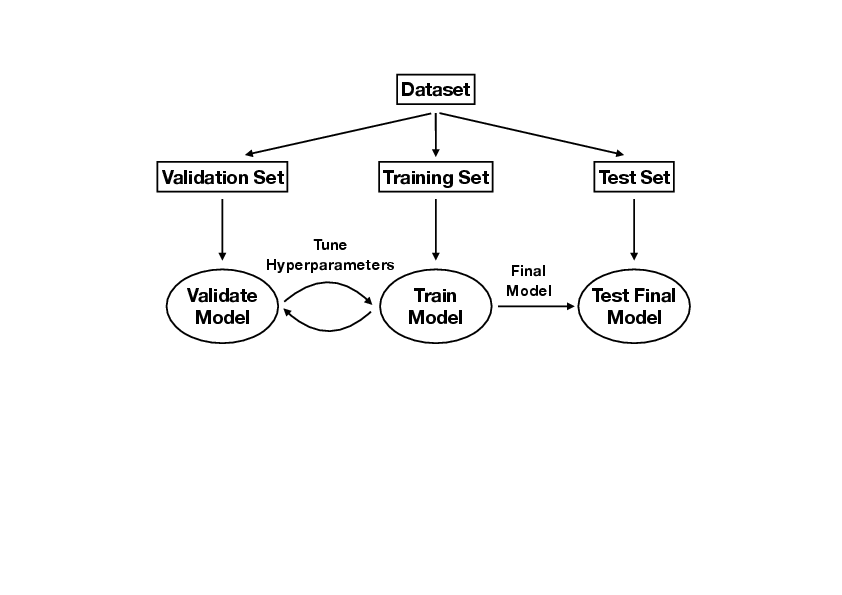
<div id="fig: ML Workflow" class="d-flex justify-content-center"> | <div id="fig: ML Workflow" class="d-flex justify-content-center"> | ||
| Line 272: | Line 272: | ||
</div> | </div> | ||
====Example==== | ====Example==== | ||
We illustrate the concepts of linear regression using a medical dataset. In the process, we will also familiarize ourselves with the standard machine learning workflow [see Fig. | We illustrate the concepts of linear regression using a medical dataset. In the process, we will also familiarize ourselves with the standard machine learning workflow [see Fig.[[#fig: ML Workflow |fig: ML Workflow]]]. For this example, we are given <math>10</math> data features, namely age, sex, body mass index, average blood pressure, and six blood serum measurements from <math>442</math> diabetes patients, and our task is train a model <math>f(\bm{x}|\bm{\beta})</math> [Eq.\eqref{eqn: Univariate Linear Model}] to predict a quantitative measure of the disease progression after one year. | ||
Recall that the final aim of a machine-learning task is not to obtain the smallest possible value for the loss function such as the RSS, but to minimise the generalisation error on unseen data [c.f. Eq. | Recall that the final aim of a machine-learning task is not to obtain the smallest possible value for the loss function such as the RSS, but to minimise the generalisation error on unseen data [c.f. Eq.\eqref{eqn: MSE Generalisation Error}]. The standard approach relies on a division of the dataset into three subsets: training set, validation set and test set. The standard workflow is summarised in Box | ||
<proc label="ML Workflow" id = "box: ML Workflow"> | |||
<ul><li> Divide the dataset into training set <math>\mathcal{T}</math>, validation set <math>\mathcal{V}</math> and test set <math>\mathcal{S}</math>. A common ratio for the split is <math>70 : 15 : 15</math>. | <ul><li> Divide the dataset into training set <math>\mathcal{T}</math>, validation set <math>\mathcal{V}</math> and test set <math>\mathcal{S}</math>. A common ratio for the split is <math>70 : 15 : 15</math>. | ||
</li> | </li> | ||
<li> Pick the hyperparameters, e.g., <math>\lambda</math> in Eq. | <li> Pick the hyperparameters, e.g., <math>\lambda</math> in Eq.\eqref{eqn: Ridge}. | ||
</li> | </li> | ||
<li> Train the model with only the training set, in other words minimize the loss function on the training set. [This corresponds to Eq. | <li> Train the model with only the training set, in other words minimize the loss function on the training set. [This corresponds to Eq.\eqref{eqn: LG RSS Solution} or \eqref{eqn: Ridge Solution} for the linear regression, where <math>\widetilde{X}</math> only contains the training set.] | ||
</li> | </li> | ||
<li> Evaluate the MSE (or any other chosen metric) on the validation set, [c.f. Eq. | <li> Evaluate the MSE (or any other chosen metric) on the validation set, [c.f. Eq.\eqref{eqn: MSE Generalisation Error}] | ||
<math display="block"> | <math display="block"> | ||
| Line 301: | Line 302: | ||
</li> | </li> | ||
</ul> | </ul> | ||
</proc> | |||
It is important to note that the test set <math>\mathcal{S}</math> was not involved in optimizing either parameters <math>\bm{\beta}</math> or the hyperparameters such as <math>\lambda</math>. | It is important to note that the test set <math>\mathcal{S}</math> was not involved in optimizing either parameters <math>\bm{\beta}</math> or the hyperparameters such as <math>\lambda</math>. | ||
Applying this procedure to the diabetes dataset<ref group="Notes" >Source: [https://www4.stat.ncsu.edu/~boos/var.select/diabetes.html https://www4.stat.ncsu.edu/ | Applying this procedure to the diabetes dataset<ref group="Notes" >Source: [https://www4.stat.ncsu.edu/~boos/var.select/diabetes.html https://www4.stat.ncsu.edu/~boos/var.select/diabetes.html]</ref>, we obtain the results in Fig.[[#fig: Regression on Diabetes Dataset |fig: Regression on Diabetes Dataset]]. We compare RSS linear regression with the ridge regression, and indeed we see that by appropriately choosing the regularisation hyperparameter <math>\lambda</math>, the generalisation error can be minimized. | ||
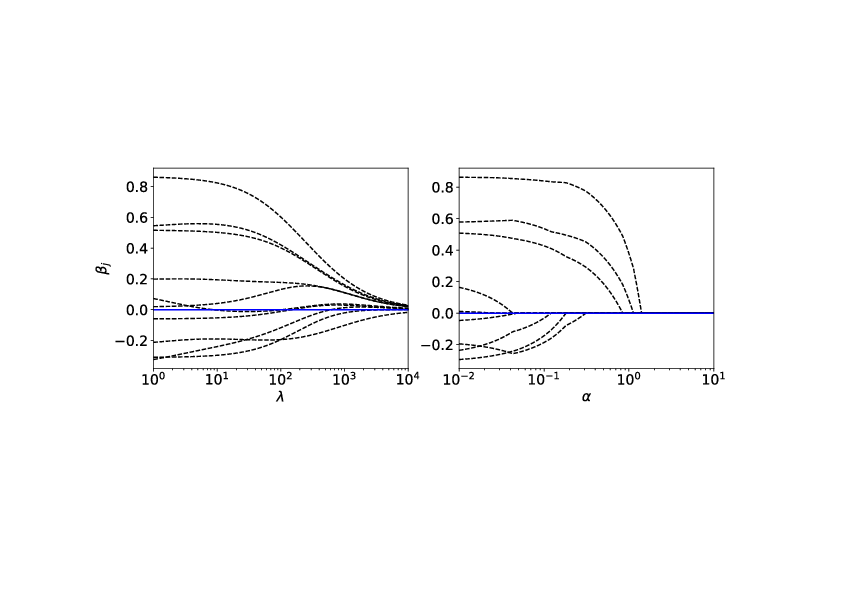
As side remark regarding the ridge regression, we can see on the left of Fig. | As side remark regarding the ridge regression, we can see on the left of Fig.[[#fig: Ridge Parameters |fig: Ridge Parameters]], that as <math>\lambda</math> increases, the magnitude of the parameters, Eq.\eqref{eqn: Ridge Solution}, <math>\hat{\bm{\beta}}_{\textrm{ridge}}</math> decreases. Consider on the other hand, a different form of regularisation, which goes by the name ''lasso regression'', where the loss function is given by | ||
<math display="block"> | <math display="block"> | ||
| Line 311: | Line 313: | ||
\end{equation} | \end{equation} | ||
</math> | </math> | ||
Despite the similarities, lasso regression has a very different behaviour as depicted on the right of Fig. | Despite the similarities, lasso regression has a very different behaviour as depicted on the right of Fig.[[#fig: Ridge Parameters |fig: Ridge Parameters]]. Notice that as <math>\alpha</math> increases some parameters actually vanish and can be ignored completely. This actually corresponds to dropping certain data features completely and can be useful if we are interested in selecting the most important features in a dataset. | ||
<div id="fig: Regression on Diabetes Dataset" class="d-flex justify-content-center"> | <div id="fig: Regression on Diabetes Dataset" class="d-flex justify-content-center"> | ||
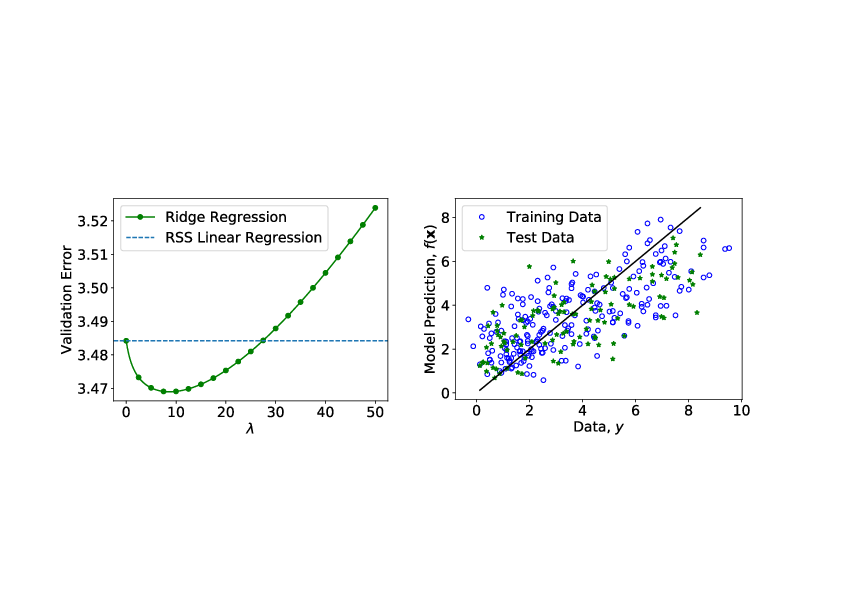
[[File:guide_7132c_diabetes_ridge_regression.png | 400px | thumb | '''Ridge Regression on Diabetes patients dataset.''' Left: Validation error versus <math>\lambda</math>. Right: Test data versus the prediction from the trained model. If | [[File:guide_7132c_diabetes_ridge_regression.png | 400px | thumb | '''Ridge Regression on Diabetes patients dataset.''' Left: Validation error versus <math>\lambda</math>. Right: Test data versus the prediction from the trained model. If | ||
| Line 349: | Line 351: | ||
</math> | </math> | ||
where <math>M</math> is called the ''margin''. | where <math>M</math> is called the ''margin''. | ||
The optimal solution <math>\hat{\bm{\beta}}</math> then maximizes this margin. Note that instead of fixing the norm of <math>\beta_{j\geq1}</math> and maximizing <math>M</math>, it is customary to minimize <math>\sum_{j=1}^n \beta_j^2</math> setting <math>M=1</math> in Eq. | The optimal solution <math>\hat{\bm{\beta}}</math> then maximizes this margin. Note that instead of fixing the norm of <math>\beta_{j\geq1}</math> and maximizing <math>M</math>, it is customary to minimize <math>\sum_{j=1}^n \beta_j^2</math> setting <math>M=1</math> in Eq.\eqref{eq:separable}. | ||
In most cases, the two classes are not completely separable. In order to still find a good classifier, we allow some of the points <math>\bm{x}_i</math> to lie within the margin or even on the wrong side of the hyperplane. For this purpose, we rewrite the optimization constraint Eq. | In most cases, the two classes are not completely separable. In order to still find a good classifier, we allow some of the points <math>\bm{x}_i</math> to lie within the margin or even on the wrong side of the hyperplane. For this purpose, we rewrite the optimization constraint Eq.\eqref{eq:separable} to | ||
<math display="block"> | <math display="block"> | ||
| Line 366: | Line 368: | ||
\end{equation} | \end{equation} | ||
</math> | </math> | ||
subject to the constraint Eq. | subject to the constraint Eq.\eqref{eq:notseparable}. Note that the second term with hyperparameter <math>C</math> acts like a regularizer, in particular a lasso regularizer. As we have seen in the example of the previous section, such a regularizer tries to set as many <math>\xi_i</math> to zero as possible. | ||
<div id="fig:svm" class="d-flex justify-content-center"> | <div id="fig:svm" class="d-flex justify-content-center"> | ||
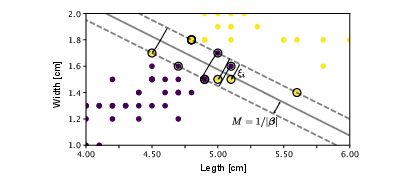
[[File:guide_7132c_SVM_overlap.png | 400px | thumb | ''' Binary classification.''' Hyperplane separating the two classes and margin <math>M</math> of the linear binary classifier. The support vectors are denoted by a circle around them. ]] | [[File:guide_7132c_SVM_overlap.png | 400px | thumb | ''' Binary classification.''' Hyperplane separating the two classes and margin <math>M</math> of the linear binary classifier. The support vectors are denoted by a circle around them. ]] | ||
| Line 396: | Line 398: | ||
\end{equation} | \end{equation} | ||
</math> | </math> | ||
subject to <math>\sum_i \alpha_i y_i =0</math> and <math>0\leq \alpha_i \leq C</math> | subject to <math>\sum_i \alpha_i y_i =0</math> and <math>0\leq \alpha_i \leq C</math><ref group="Notes" >Note that the constraints for the minimization are not equalities, but actually inequalities. A solution thus has to fulfil the additional Karush-Kuhn-Tucker constraints | ||
<math display="block"> | <math display="block"> | ||
| Line 406: | Line 408: | ||
</math> | </math> | ||
</ref>. | </ref>. | ||
Using Eq. | Using Eq.\eqref{eq:svm_beta}, we can reexpress <math>\beta_j</math> to find | ||
<math display="block"> | <math display="block"> | ||
| Line 414: | Line 416: | ||
\end{equation} | \end{equation} | ||
</math> | </math> | ||
where the sum only runs over the points <math>\bm{x}_i</math>, which lie within the margin, as all other points have <math>\alpha_i\equiv0</math> [see Eq. | where the sum only runs over the points <math>\bm{x}_i</math>, which lie within the margin, as all other points have <math>\alpha_i\equiv0</math> [see Eq.\eqref{eq:firstKKT}]. These points are thus called the ''support vectors'' and are denoted in Fig.[[#fig:svm |fig:svm]] with a circle around them. Finally, note that we can use Eq.\eqref{eq:firstKKT} again to find <math>\beta_0</math>. | ||
'''The Kernel trick and support vector machines''' | |||
We have seen in our discussion of PCA that most data is not separable linearly. However, we have also seen how the kernel trick can help us in such situations. In particular, we have seen how a non-linear function <math>\bm{\Phi}(\bm{x})</math>, which we first apply to the data <math>\bm{x}</math>, can help us separate data that is not linearly separable. Importantly, we never actually use the non-linear function <math>\bm{\Phi}(\bm{x})</math>, but only the kernel. | We have seen in our discussion of PCA that most data is not separable linearly. However, we have also seen how the kernel trick can help us in such situations. In particular, we have seen how a non-linear function <math>\bm{\Phi}(\bm{x})</math>, which we first apply to the data <math>\bm{x}</math>, can help us separate data that is not linearly separable. Importantly, we never actually use the non-linear function <math>\bm{\Phi}(\bm{x})</math>, but only the kernel. | ||
Looking at the dual optimization problem Eq. | Looking at the dual optimization problem Eq.\eqref{eq:svm_dual} and the resulting classifier Eq.\eqref{eq:svm_f}, we see that, as in the case of Kernel PCA, only the kernel <math>K(\bm{x}, \bm{y}) = \bm{\Phi}(\bm{x})^T\bm{\Phi}(\bm{y})</math> enters, simplifying the problem. This non-linear extension of the binary classifier is called a ''support vector machine''. | ||
====More than two classes: logistic regression==== | ====More than two classes: logistic regression==== | ||
| Line 446: | Line 449: | ||
where <math>e^{(y)}_l = 1</math> if <math>l = y</math> and zero for all other <math>l=1,\ldots, p</math>. A main advantage of this encoding is that we are not forced to choose a potentially biasing ordering of the classes as we would when arranging them along the ray of integers. | where <math>e^{(y)}_l = 1</math> if <math>l = y</math> and zero for all other <math>l=1,\ldots, p</math>. A main advantage of this encoding is that we are not forced to choose a potentially biasing ordering of the classes as we would when arranging them along the ray of integers. | ||
A linear approach to this problem then again mirrors the case for linear regression. | A linear approach to this problem then again mirrors the case for linear regression. | ||
We fit a multi-variate linear model, Eq. | We fit a multi-variate linear model, Eq.\eqref{eqn: Multivariate Linear Model}, to the one-hot encoded dataset \allowbreak<math>\lbrace(\bm{x}_{1}, \bm{e}^{(y_1)}), \dots, (\bm{x}_{m}, \bm{e}^{(y_m)})\rbrace</math>. By minimising the RSS, Eq.\eqref{eqn: RSS}, we obtain the solution | ||
<math display="block"> | <math display="block"> | ||
| Line 469: | Line 472: | ||
</math> | </math> | ||
Importantly, the output of the softmax function is a probability <math>P(y = k|\bm{x})</math>, since <math>\sum_k F_k(\bm{x}|\hat{\beta}) = 1</math>. | Importantly, the output of the softmax function is a probability <math>P(y = k|\bm{x})</math>, since <math>\sum_k F_k(\bm{x}|\hat{\beta}) = 1</math>. | ||
This extended linear model is referred to as ''logistic regression'' | This extended linear model is referred to as ''logistic regression''<ref group="Notes" >Note that the softmax function for two classes is the logistic function.</ref>. | ||
The current linear approach based on classification of one-hot encoded data generally works poorly when there are more than two classes. We will see in the next chapter that relatively straightforward non-linear extensions of this approach can lead to much better results. | The current linear approach based on classification of one-hot encoded data generally works poorly when there are more than two classes. We will see in the next chapter that relatively straightforward non-linear extensions of this approach can lead to much better results. | ||
==General references== | ==General references== | ||
Revision as of 10:22, 17 May 2024
\label{sec: linear methods for supervised learning} Supervised learning is the term for a machine learning task, where we are given a dataset consisting of input-output pairs [math]\lbrace(\bm{x}_{1}, y_{1}), \dots, (\bm{x}_{m}, y_{m})\rbrace[/math] and our task is to "learn" a function which maps input to output [math]f: \bm{x} \mapsto y[/math]. Here we chose a vector-valued input [math]\bm{x}[/math] and only a single real number as output [math]y[/math], but in principle also the output can be vector valued. The output data that we have is called the ground truth and sometimes also referred to as “labels” of the input. In contrast to supervised learning, all algorithms presented so far were unsupervised, because they just relied on input-data, without any ground truth or output data.
Within the scope of supervised learning, there are two main types of tasks: Classification and Regression. In a classification task, our output [math]y[/math] is a discrete variable corresponding to a classification category. An example of such a task would be to distinguish stars with a planetary system (exoplanets) from those without given time series of images of such objects. On the other hand, in a regression problem, the output [math]y[/math] is a continuous number or vector. For example predicting the quantity of rainfall based on meteorological data from the previous days. In this section, we first familiarize ourselves with linear methods for achieving these tasks. Neural networks, in contrast, are a non-linear method for supervised classification and regression tasks.
Linear regression
Linear regression, as the name suggests, simply means to fit a linear model to a dataset. Consider a dataset consisting of input-output pairs [math]\lbrace(\bm{x}_{1}, y_{1}), \dots, (\bm{x}_{m}, y_{m})\rbrace[/math], where the inputs are [math]n[/math]-component vectors [math]\boldsymbol{x}^{T} = (x_1, x_2, \dots , x_n)[/math] and the output [math]y[/math] is a real-valued number. The linear model then takes the form
or in matrix notation
where [math]\bm{\tilde{x}}^{T} = (1, x_1, x_2, \dots , x_n)[/math] and [math]\bm{\beta} = (\beta_0, \dots, \beta_n)^{T}[/math] are [math](n+1)[/math] dimensional row vectors. The aim then is to find parameters [math]\hat{\bm{\beta}}[/math] such that [math]f(\bm{x}|\hat{\bm{\beta}})[/math] is a good estimator for the output value [math]y[/math]. In order to quantify what it means to be a “good” estimator, one need to specify a real-valued loss function [math]L(\bm{\beta})[/math], sometimes also called a cost function. The good set of parameters [math]\hat{\bm{\beta}}[/math] is then the minimizer of this loss function
There are many, inequivalent, choices for this loss function. For our purpose, we choose the loss function to be residual sum of squares (RSS) defined as
where the sum runs over the [math]m[/math] samples of the dataset. This loss function is sometimes also called the L2-loss and can be seen as a measure of the distance between the output values from the dataset [math]y_i[/math] and the corresponding predictions [math]f(\bm{x}_i|\bm{\beta})[/math]. It is convenient to define the [math]m[/math] by [math](n+1)[/math] data matrix [math]\widetilde{X}[/math], each row of which corresponds to an input sample [math]\bm{\tilde{x}}^{T}_{i}[/math], as well as the output vector [math]\bm{Y}^{T} = (y_{1}, \dots, y_{m})[/math]. With this notation, Eq.\eqref{eqn: RSS} can be expressed succinctly as a matrix equation
The minimum of [math]\textrm{RSS}(\bm{\beta})[/math] can be easily solved by considering the partial derivatives with respect to [math]\bm{\beta}[/math], i.e.,
At the minimum, [math]\frac{\partial \textrm{RSS}}{\partial \bm{\beta}} = 0[/math] and [math]\frac{\partial^{2} \textrm{RSS}}{\partial \bm{\beta}\partial \bm{\beta}^{T}}[/math] is positive-definite. Assuming [math] \widetilde{X}^{T}\widetilde{X}[/math] is full-rank and hence invertible, we can obtain the solution [math]\hat{\bm{\beta}}[/math] as
If [math] \widetilde{X}^{T}\widetilde{X}[/math] is not full-rank, which can happen if certain data features are perfectly correlated (e.g., [math]x_1 = 2x_3[/math]), the solution to [math]\widetilde{X}^{T}\widetilde{X}\bm{\beta} = \widetilde{X}^{T}\bm{Y}[/math] can still be found, but it would not be unique. Note that the RSS is not the only possible choice for the loss function and a different choice would lead to a different solution. What we have done so far is uni-variate linear regression, that is linear regression where the output [math]y[/math] is a single real-valued number. The generalisation to the multi-variate case, where the output is a [math]p[/math]-component vector [math]\bm{y}^{T} = (y_1, \dots y_p)[/math], is straightforward. The model takes the form
where the parameters [math]\beta_{jk}[/math] now have an additional index [math]k = 1, \dots, p[/math]. Considering the parameters [math]\beta[/math] as a [math](n+1)[/math] by [math]p[/math] matrix, we can show that the solution takes the same form as before [Eq.\eqref{eqn: LG RSS Solution}] with [math]Y[/math] as a [math]m[/math] by [math]p[/math] output matrix.
Statistical analysis
Let us stop here and evaluate the quality of the method we have just introduced. At the same time, we will take the opportunity to introduce some statistics notions, which will be useful throughout the book. Up to now, we have made no assumptions about the dataset we are given, we simply stated that it consisted of input-output pairs, [math]\{(\bm{x}_{1}, y_{1}), \dots,[/math] [math](\bm{x}_{m}, y_{m})\}[/math]. In order to assess the accuracy of our model in a mathematically clean way, we have to make an additional assumption. The output data [math]y_1\ldots, y_m[/math] may arise from some measurement or observation. Then, each of these values will generically be subject to errors [math]\epsilon_1,\cdots, \epsilon_m[/math] by which the values deviate from the “true” output without errors,
We assume that this error [math]\epsilon[/math] is a Gaussian random variable with mean [math]\mu = 0[/math] and variance [math]\sigma^2[/math], which we denote by [math]\epsilon \sim \mathcal{N}(0, \sigma^2)[/math]. Assuming that a linear model in Eq.\eqref{eqn: Univariate Linear Model} is a suitable model for our dataset, we are interested in the following question: How does our solution [math]\hat{\bm{\beta}}[/math] as given in Eq.\eqref{eqn: LG RSS Solution} compare with the true solution [math]\bm{\beta}^{\textrm{true}}[/math] which obeys
In order to make statistical statements about this question, we have to imagine that we can fix the inputs [math]\bm{x}_{i}[/math] of our dataset and repeatedly draw samples for our outputs [math]y_i[/math]. Each time we will obtain a different value for [math]y_i[/math] following Eq.\eqref{eqn: True Linear}, in other words the [math]\epsilon_i[/math] are uncorrelated random numbers. This allows us to formalise the notion of an expectation value [math]E(\cdots)[/math] as the average over an infinite number of draws. For each draw, we obtain a new dataset, which differs from the other ones by the values of the outputs [math]y_i[/math]. With each of these datasets, we obtain a different solution [math]\hat{\bm{\beta}}[/math] as given by Eq.\eqref{eqn: LG RSS Solution}. The expectation value [math]E(\hat{\bm{\beta}})[/math] is then simply the average value we obtained across an infinite number of datasets. The deviation of this average value from the “true” value given perfect data is called the bias of the model,
For the linear regression we study here, the bias is exactly zero, because
where the second line follows because [math]E(\bm{\epsilon}) = \bm{0}[/math] and [math](\widetilde{X}^{T}\widetilde{X})^{-1} \widetilde{X}^{T} \bm{Y}^{\textrm{true}} = \bm{\beta}^{\textrm{true}}[/math]. Equation\eqref{eqn: LG RSS Unbiased} implies linear regression is unbiased. Note that other machine learning algorithms will in general be biased. What about the standard error or uncertainty of our solution? This information is contained in the covariance matrix
The covariance matrix can be computed for the case of linear regression using the solution in Eq.\eqref{eqn: LG RSS Solution}, the expectation value in Eq.\eqref{eqn: LG RSS Unbiased} and the assumption in Eq.\eqref{eqn: True Linear} that [math]Y = Y^{\textrm{true}} + \bm{\epsilon}[/math] yielding
This expression can be simplified by using the fact that our input matrices [math]\widetilde{X}[/math] are independent of the draw such that
Here, the second line follows from the fact that different samples are uncorrelated, which implies that [math]E(\bm{\epsilon} \bm{\epsilon}^{T}) = \sigma^2 I[/math] with [math]I[/math] the identity matrix. The diagonal elements of [math]\sigma^2 (\widetilde{X}^{T}\widetilde{X})^{-1}[/math] then correspond to the variance
of the individual parameters [math]\beta_i[/math]. The standard error or uncertainty is then [math]\sqrt{\textrm{Var}(\hat{\beta}_{i})}[/math]. There is one more missing element: we have not explained how to obtain the variances [math]\sigma^2[/math] of the outputs [math]y[/math]. In an actual machine learning task, we would not know anything about the true relation, as given in Eq.\eqref{eqn: True Linear}, governing our dataset. The only information we have access to is a single dataset. Therefore, we have to estimate the variance using the samples in our dataset, which is given by
where [math]y_i[/math] are the output values from our dataset and [math]f(\bm{x}_i|\hat{\bm{\beta}})[/math] is the corresponding prediction. Note that we normalized the above expression by [math](m - n - 1)[/math] instead of [math]m[/math] to ensure that [math]E(\hat{\sigma}^2) = \sigma^2[/math], meaning that [math]\hat{\sigma}^2[/math] is an unbiased estimator of [math]\sigma^2[/math]. Our ultimate goal is not simply to fit a model to the dataset. We want our model to generalize to inputs not within the dataset. To assess how well this is achieved, let us consider the prediction [math]\tilde{\bm{a}}^{T} \hat{\bm{\beta}}[/math] on a new random input-output pair [math](\bm{a},y_{0})[/math]. The output is again subject to an error [math]y_{0} = \tilde{\bm{a}}^{T}\bm{\beta}^{\textrm{true}} + \epsilon[/math]. In order to compute the expected error of the prediction, we compute the expectation value of the loss function over these previously unseen data. This is also known as the test or generalization error . For the square-distance loss function, this is the mean square error (MSE)
There are three terms in the expression. The first term is the irreducible or intrinsic uncertainty of the dataset. The second term represents the bias and the third term is the variance of the model. For RSS linear regression, the estimate is unbiased so that
Based on the assumption that the dataset indeed derives from a linear model as given by Eq.\eqref{eqn: True Linear} with a Gaussian error, it can be shown that the RSS solution, Eq.\eqref{eqn: LG RSS Solution}, gives the minimum error among all unbiased linear estimators, Eq.\eqref{eqn: Univariate Linear Model}. This is known as the Gauss-Markov theorem. This completes our error analysis of the method.
Regularization and the bias-variance tradeoff
Although the RSS solution has the minimum error among unbiased linear estimators, the expression for the generalisation error, Eq.\eqref{eqn: MSE Generalisation Error}, suggests that we can actually still reduce the error by sacrificing some bias in our estimate.
A possible way to reduce generalisation error is actually to drop some data features. From the [math]n[/math] data features [math]\lbrace x_{1}, \dots x_{n} \rbrace[/math], we can pick a reduced set [math]\mathcal{M}[/math]. For example, we can choose [math]\mathcal{M} = \lbrace x_{1}, x_{3}, x_{7} \rbrace[/math], and define our new linear model as
This is equivalent to fixing some parameters to zero, i.e., [math]\beta_k = 0[/math] if [math]x_{k} \notin \mathcal{M}[/math]. Minimizing the RSS with this constraint results in a biased estimator but the reduction in model variance can sometimes help to reduce the overall generalisation error. For a small number of features [math]n \sim 20[/math], one can search exhaustively for the best subset of features that minimises the error, but beyond that the search becomes computationally unfeasible. A common alternative is called ridge regression. In this method, we consider the same linear model given in Eq.\eqref{eqn: Univariate Linear Model} but with a modified loss function
where [math]\lambda \gt 0[/math] is a positive parameter. This is almost the same as the RSS apart from the term proportional to [math]\lambda[/math] [c.f. Eq.\eqref{eqn: RSS}]. The effect of this new term is to penalize large parameters [math]\beta_j[/math] and bias the model towards smaller absolute values. The parameter [math]\lambda[/math] is an example of a hyper-parameter\index{hyper-parameter}, which is kept fixed during the training. On fixing [math]\lambda[/math] and minimising the loss function, we obtain the solution
from which we can see that as [math]\lambda \rightarrow \infty[/math], [math]\hat{\bm{\beta}}_{\textrm{ridge}} \rightarrow \bm{0}[/math]. By computing the bias and variance,
it is also obvious that increasing [math]\lambda[/math] increases the bias, while reducing the variance. This is the tradeoff between bias and variance. By appropriately choosing [math]\lambda[/math] it is possible that generalisation error can be reduced. We will introduce in the next section a common strategy how to find the optimal value for [math]\lambda[/math]. The techniques presented here to reduce the generalization error, namely dropping of features and biasing the model to small parameters, are part of a large class of methods known as regularization. Comparing the two methods, we can see a similarity. Both methods actually reduce the complexity of our model. In the former, some parameters are set to zero, while in the latter, there is a constraint which effectively reduces the magnitude of all parameters. A less complex model has a smaller variance but larger bias. By balancing these competing effects, generalisation can be improved, as illustrated schematically in Fig.fig: Bias-Variance Tradeoff. In the next chapter, we will see that these techniques are useful beyond applications to linear methods. We illustrate the different concepts in the following example.
Example
We illustrate the concepts of linear regression using a medical dataset. In the process, we will also familiarize ourselves with the standard machine learning workflow [see Fig.fig: ML Workflow]. For this example, we are given [math]10[/math] data features, namely age, sex, body mass index, average blood pressure, and six blood serum measurements from [math]442[/math] diabetes patients, and our task is train a model [math]f(\bm{x}|\bm{\beta})[/math] [Eq.\eqref{eqn: Univariate Linear Model}] to predict a quantitative measure of the disease progression after one year. Recall that the final aim of a machine-learning task is not to obtain the smallest possible value for the loss function such as the RSS, but to minimise the generalisation error on unseen data [c.f. Eq.\eqref{eqn: MSE Generalisation Error}]. The standard approach relies on a division of the dataset into three subsets: training set, validation set and test set. The standard workflow is summarised in Box
- Divide the dataset into training set [math]\mathcal{T}[/math], validation set [math]\mathcal{V}[/math] and test set [math]\mathcal{S}[/math]. A common ratio for the split is [math]70 : 15 : 15[/math].
- Pick the hyperparameters, e.g., [math]\lambda[/math] in Eq.\eqref{eqn: Ridge}.
- Train the model with only the training set, in other words minimize the loss function on the training set. [This corresponds to Eq.\eqref{eqn: LG RSS Solution} or \eqref{eqn: Ridge Solution} for the linear regression, where [math]\widetilde{X}[/math] only contains the training set.]
- Evaluate the MSE (or any other chosen metric) on the validation set, [c.f. Eq.\eqref{eqn: MSE Generalisation Error}]
[[math]] \begin{equation} \textrm{MSE}_{\textrm{validation}}(\hat{\bm{\beta}}) = \frac{1}{|\mathcal{V}|}\sum_{j\in\mathcal{V}} (y_j - f(\bm{x}_j|\hat{\bm{\beta}}))^2. \end{equation} [[/math]]This is known as the validation error.
- Pick a different value for the hyperparameters and repeat steps [math]3[/math] and [math]4[/math], until validation error is minimized.
- Evaluate the final model on the test set
[[math]] \begin{equation} \textrm{MSE}_{\textrm{test}}(\hat{\bm{\beta}}) = \frac{1}{|\mathcal{S}|}\sum_{j\in\mathcal{S}} (y_j - f(\bm{x}_j|\hat{\bm{\beta}}))^2. \end{equation} [[/math]]
It is important to note that the test set [math]\mathcal{S}[/math] was not involved in optimizing either parameters [math]\bm{\beta}[/math] or the hyperparameters such as [math]\lambda[/math]. Applying this procedure to the diabetes dataset[Notes 1], we obtain the results in Fig.fig: Regression on Diabetes Dataset. We compare RSS linear regression with the ridge regression, and indeed we see that by appropriately choosing the regularisation hyperparameter [math]\lambda[/math], the generalisation error can be minimized. As side remark regarding the ridge regression, we can see on the left of Fig.fig: Ridge Parameters, that as [math]\lambda[/math] increases, the magnitude of the parameters, Eq.\eqref{eqn: Ridge Solution}, [math]\hat{\bm{\beta}}_{\textrm{ridge}}[/math] decreases. Consider on the other hand, a different form of regularisation, which goes by the name lasso regression, where the loss function is given by
Despite the similarities, lasso regression has a very different behaviour as depicted on the right of Fig.fig: Ridge Parameters. Notice that as [math]\alpha[/math] increases some parameters actually vanish and can be ignored completely. This actually corresponds to dropping certain data features completely and can be useful if we are interested in selecting the most important features in a dataset.
Linear classifiers and their extensions
Binary classification and support vector machines
In a classification problem, the aim is to categorize the inputs into one of a finite set of classes. Formulated as a supervised learning task, the dataset again consists of input-output pairs, i.e. [math]\lbrace(\bm{x}_{1}, y_{1}), \dots, (\bm{x}_{m}, y_{m})\rbrace[/math] with [math]\bm{x}\in \mathbb{R}^n[/math]. However, unlike regression problems, the output [math]y[/math] is a discrete integer number representing one of the classes. In a binary classification problem, in other words a problem with only two classes, it is natural to choose [math]y\in\{-1, 1\}[/math]. We have introduced linear regression in the previous section as a method for supervised learning when the output is a real number. Here, we will see how we can use the same model for a binary classification task. If we look at the regression problem, we first note that geometrically
defines a hyperplane perpendicular to the vector with elements [math]\beta_{j\geq1}[/math]. If we fix the length [math]\sum_{j=1}^n \beta_j^2=1[/math], then [math]f(\bm{x}|\bm{\beta})[/math] measures the (signed) distance of [math]\bm{x}[/math] to the hyperplane with a sign depending on which side of the plane the point [math]\bm{x}_i[/math] lies. To use this model as a classifier, we thus define
which yields [math]\{+1, -1\}[/math]. If the two classes are (completely) linearly separable, then the goal of the classification is to find a hyperplane that separates the two classes in feature space. Specifically, we look for parameters [math]\bm{\beta}[/math], such that
where [math]M[/math] is called the margin. The optimal solution [math]\hat{\bm{\beta}}[/math] then maximizes this margin. Note that instead of fixing the norm of [math]\beta_{j\geq1}[/math] and maximizing [math]M[/math], it is customary to minimize [math]\sum_{j=1}^n \beta_j^2[/math] setting [math]M=1[/math] in Eq.\eqref{eq:separable}. In most cases, the two classes are not completely separable. In order to still find a good classifier, we allow some of the points [math]\bm{x}_i[/math] to lie within the margin or even on the wrong side of the hyperplane. For this purpose, we rewrite the optimization constraint Eq.\eqref{eq:separable} to
We can now define the optimization problem as finding
subject to the constraint Eq.\eqref{eq:notseparable}. Note that the second term with hyperparameter [math]C[/math] acts like a regularizer, in particular a lasso regularizer. As we have seen in the example of the previous section, such a regularizer tries to set as many [math]\xi_i[/math] to zero as possible.
We can solve this constrained minimization problem by introducing Lagrange multipliers [math]\alpha_i[/math] and [math]\mu_i[/math] and solving
which yields the conditions
It is numerically simpler to solve the dual problem
subject to [math]\sum_i \alpha_i y_i =0[/math] and [math]0\leq \alpha_i \leq C[/math][Notes 2]. Using Eq.\eqref{eq:svm_beta}, we can reexpress [math]\beta_j[/math] to find
where the sum only runs over the points [math]\bm{x}_i[/math], which lie within the margin, as all other points have [math]\alpha_i\equiv0[/math] [see Eq.\eqref{eq:firstKKT}]. These points are thus called the support vectors and are denoted in Fig.fig:svm with a circle around them. Finally, note that we can use Eq.\eqref{eq:firstKKT} again to find [math]\beta_0[/math].
The Kernel trick and support vector machines
We have seen in our discussion of PCA that most data is not separable linearly. However, we have also seen how the kernel trick can help us in such situations. In particular, we have seen how a non-linear function [math]\bm{\Phi}(\bm{x})[/math], which we first apply to the data [math]\bm{x}[/math], can help us separate data that is not linearly separable. Importantly, we never actually use the non-linear function [math]\bm{\Phi}(\bm{x})[/math], but only the kernel. Looking at the dual optimization problem Eq.\eqref{eq:svm_dual} and the resulting classifier Eq.\eqref{eq:svm_f}, we see that, as in the case of Kernel PCA, only the kernel [math]K(\bm{x}, \bm{y}) = \bm{\Phi}(\bm{x})^T\bm{\Phi}(\bm{y})[/math] enters, simplifying the problem. This non-linear extension of the binary classifier is called a support vector machine.
More than two classes: logistic regression
In the following, we are interested in the case of [math]p[/math] classes with [math]p \gt 2[/math]. After the previous discussion, it seems natural for the output to take the integer values [math]y = 1, \dots, p[/math]. However, it turns out to be helpful to use a different, so-called one-hot encoding\index{one-hot encoding}. In this encoding, the output [math]y[/math] is instead represented by the [math]p[/math]-dimensional unit vector in [math]y[/math] direction [math]\bm{e}^{(y)}[/math],
where [math]e^{(y)}_l = 1[/math] if [math]l = y[/math] and zero for all other [math]l=1,\ldots, p[/math]. A main advantage of this encoding is that we are not forced to choose a potentially biasing ordering of the classes as we would when arranging them along the ray of integers. A linear approach to this problem then again mirrors the case for linear regression. We fit a multi-variate linear model, Eq.\eqref{eqn: Multivariate Linear Model}, to the one-hot encoded dataset \allowbreak[math]\lbrace(\bm{x}_{1}, \bm{e}^{(y_1)}), \dots, (\bm{x}_{m}, \bm{e}^{(y_m)})\rbrace[/math]. By minimising the RSS, Eq.\eqref{eqn: RSS}, we obtain the solution
where [math]Y[/math] is the [math]m[/math] by [math]p[/math] output matrix. The prediction given an input [math]\bm{x}[/math] is then a [math]p[/math]-dimensional vector [math]\bm{f}(\bm{x}|\hat{\beta}) = \tilde{\bm{x}}^{T} \hat{\beta}[/math]. On a generic input [math]\bm{x}[/math], it is obvious that the components of this prediction vector would be real valued, rather than being one of the one-hot basis vectors. To obtain a class prediction [math]F(\bm{x}|\hat{\beta}) = 1, \dots, p[/math], we simply take the index of the largest component of that vector, i.e.,
The [math]\textrm{argmax}[/math] function is a non-linear function and is a first example of what is referred to as activation function. For numerical minimization, it is better to use a smooth activation function. Such an activation function is given by the softmax function
Importantly, the output of the softmax function is a probability [math]P(y = k|\bm{x})[/math], since [math]\sum_k F_k(\bm{x}|\hat{\beta}) = 1[/math]. This extended linear model is referred to as logistic regression[Notes 3]. The current linear approach based on classification of one-hot encoded data generally works poorly when there are more than two classes. We will see in the next chapter that relatively straightforward non-linear extensions of this approach can lead to much better results.
General references
Neupert, Titus; Fischer, Mark H; Greplova, Eliska; Choo, Kenny; Denner, M. Michael (2022). "Introduction to Machine Learning for the Sciences". arXiv:2102.04883 [physics.comp-ph].
Notes
- Source: https://www4.stat.ncsu.edu/~boos/var.select/diabetes.html
- Note that the constraints for the minimization are not equalities, but actually inequalities. A solution thus has to fulfil the additional Karush-Kuhn-Tucker constraints
[[math]] \begin{eqnarray} \alpha_i [y_i \tilde{\bm{x}}_i^T\bm{\beta} - (1-\xi_i)]&=&0,\label{eq:firstKKT}\\ \mu_i\xi_i &=& 0,\\ y_i \tilde{\bm{x}}_i^T\bm{\beta} - (1-\xi_i)& \gt & 0. \end{eqnarray} [[/math]]
- Note that the softmax function for two classes is the logistic function.