Bayesian regression
[math] \require{textmacros} \def \bbeta {\bf \beta} \def\fat#1{\mbox{\boldmath$#1$}} \def\reminder#1{\marginpar{\rule[0pt]{1mm}{11pt}}\textbf{#1}} \def\SSigma{\bf \Sigma} \def\ttheta{\bf \theta} \def\aalpha{\bf \alpha} \def\bbeta{\bf \beta} \def\ddelta{\bf \delta} \def\eeta{\bf \eta} \def\llambda{\bf \lambda} \def\ggamma{\bf \gamma} \def\nnu{\bf \nu} \def\vvarepsilon{\bf \varepsilon} \def\mmu{\bf \mu} \def\nnu{\bf \nu} \def\ttau{\bf \tau} \def\SSigma{\bf \Sigma} \def\TTheta{\bf \Theta} \def\XXi{\bf \Xi} \def\PPi{\bf \Pi} \def\GGamma{\bf \Gamma} \def\DDelta{\bf \Delta} \def\ssigma{\bf \sigma} \def\UUpsilon{\bf \Upsilon} \def\PPsi{\bf \Psi} \def\PPhi{\bf \Phi} \def\LLambda{\bf \Lambda} \def\OOmega{\bf \Omega} [/math]
The ridge regression estimator is equivalent to a Bayesian regression estimator. On one hand this equivalence provides another interpretation of the ridge regression estimator. But it also shows that within the Bayesian framework the high-dimensionality of the data need not frustrate the numerical evaluation of the estimator. In addition, the framework provides ways to quantify the consequences of high-dimensionality on the uncertainty of the estimator. Within this chapter focus is on the equivalence of Bayesian and ridge regression. In this particular case, the connection is immediate from the analytic formulation of the Bayesian regression estimator. After this has been presented, it is shown how this estimator may also be obtained by means of sampling. The relevance of the sampling for the evaluation of the estimator and its uncertainty becomes apparent in subsequent chapters when discussing other penalized estimators for which the connection cannot be captured in analytic form.
A minimum of prior knowledge on Bayesian statistics
The schism between Bayesian and frequentist statistics centers on the interpretation of the concept of probability. A frequentist views the probability of an event as the limiting frequency of observing the event among a large number of trials. In contrast, a Bayesian considers it to be a measure of believe (in the occurence) of the event. The difference between the two interpretations becomes clear when considering events that can occur only once (or a small number of times). A Bayesian would happily discuss the probability of this (possibly hypothetical) event happening, which would be meaningless to a frequentist.
What are the consequences of this schism for the current purpose of estimating a regression model? Exponents from both paradigms assume a statistical model, e.g. here the linear regression model, to describe the data generating mechanism. But a frequentist treats the parameters as platonic quantities for which a true value exists that is to be estimated from the data. A Bayesian, however, first formalizes his/her current believe/knowledge on the parameters in the form of probability distributions. This is referred to as the prior -- to the experiment -- distribution. The parameters are thus random variables and their distributions are to be interpreted as reflecting the (relative) likelihood of the parameters' values. From the Bayesian point it then makes sense to talk about the random behaviour of the parameter, e.g., what is probability of that the parameter is larger than a particular value. In the frequentist context one may ask this of the estimator, but not of the parameter. The parameter either is or is not larger than this value as a platonic quantity is not governed by a chance process. Then, having specified his/her believe on the parameters, a Bayesian uses the data to update this believe of the parameters of the statistical model, which again is a distribution and called the posterior -- to the experiment -- distribution.
To illustrate this process of updating assume that the data [math]Y_1, \ldots, Y_n \sim \{0, 1 \}[/math] are assumed to be independently and identically drawn from a Bernouilli distribution with parameter [math]\theta = P(Y_i = 1)[/math]. The likelihood of these data for a given choice of the parameter [math]\theta[/math] then is: [math]L(\mathbf{Y} = \mathbf{y}; \theta) = P(\mathbf{Y} = \mathbf{y} \, | \, \theta) = P(Y_1 = y_1, \ldots, Y_n = y_n \, | \, \theta) = \prod_{i=1}^n P(Y_i = y_i \, | \, \theta) = \prod_{i=1}^n \theta^{y_i} (1-\theta)^{1-y_i}[/math]. A Bayesian now needs to specificy the prior distribution, denoted [math]\pi_{\theta}(\cdot)[/math], on the parameter [math]\theta[/math]. A common choice in this case would be a beta-distribution: [math]\theta \sim \mathcal{B}(\alpha, \beta)[/math]. The posterior distribution is now arrived at by the use of Bayes' rule:
The posterior distribution thus contains all knowledge on the parameter. As the denomenator -- referred to as the distribution's normalizing constant -- in the expression of the posterior distribution does not involve the parameter [math]\theta[/math], one often writes [math]\pi ( \theta \, | \, \mathbf{Y} = \mathbf{y}) \propto P(\mathbf{Y} = \mathbf{y} \, | \, \theta) \, \pi (\theta)[/math], thus specifying the (relative) density of the posterior distribution of [math]\theta[/math] as ‘likelihood [math]\times[/math] prior’.
While the posterior distribution is all one wishes to know, often a point estimate is required. A point estimate of [math]\theta[/math] can be obtained from the posterior by taking the mean or the mode. Formally, the Bayesian point estimator of a parameter [math]\ttheta[/math] is defined as the estimator that minimizes the Bayes risk over a prior distribution of the parameter [math]\theta[/math]. The Bayes risk is defined as [math]\int_{\theta} \mathbb{E} [(\hat{\theta} - \theta)^2] \pi_{\theta}(\theta; \alpha) d\theta[/math], where [math]\pi_{\theta}(\theta; \alpha)[/math] is the prior distribution of [math]\theta[/math] with hyperparameter [math]\alpha[/math]. It is thus a weighted average of the Mean Squared Error, with weights specified through the prior. The Bayes risk is minimized by the posterior mean:
(cf., e.g., [1]). The Bayesian point estimator of [math]\theta[/math] yields the smallest possible expected MSE, under the assumption of the employed prior. This estimator thus depends on the likelihood and the prior, and a different prior yields a different estimator.
A point estimator is preferably accompanied by a quantification of its uncertainty. Within the Bayesian context this is done by so-called credible intervals or regions, the Bayesian equivalent of confidence intervals or regions. A Bayesian credible interval encompass a certain percentage of the probability mass of the posterior. For instance, a subset of the parameter space [math]\mathcal{C}[/math] forms an [math]100 (1-\alpha) \%[/math] credible interval if [math]\int_{\mathcal{C}} \pi(\theta \, | \, \mbox{data}) \, d \theta = 1- \alpha[/math].
In the example above it is important to note the role of the prior in the updating of the knowledge on [math]\theta[/math]. First, when the prior distribution is uniform on the unit interval, the mode of the posterior coincides with the maximum likelihood estimator. Furthermore, when [math]n[/math] is large the influence of the prior in the posterior is negligible. It is therefore that for large sample sizes frequentist and Bayesian analyses tend to produce similar results. However, when [math]n[/math] is small, the prior's contribution to the posterior distribution cannot be neglected. The choice of the prior is then crucial as it determines the shape of the posterior distribution and, consequently, the posterior estimates. It is this strong dependence of the posterior on the arbitrary (?) choice of the prior that causes unease with a frequentist (leading some to accuse Bayesians of subjectivity). In the high-dimensional setting the sample size is usually small, especially in relation to the parameter dimension, and the choice of the prior distribution then matters. Interest here is not in resolving the frequentist's unease, but in identifying or illustrating the effect of the choice of the prior distribution on the parameter estimates.
Relation to ridge regression
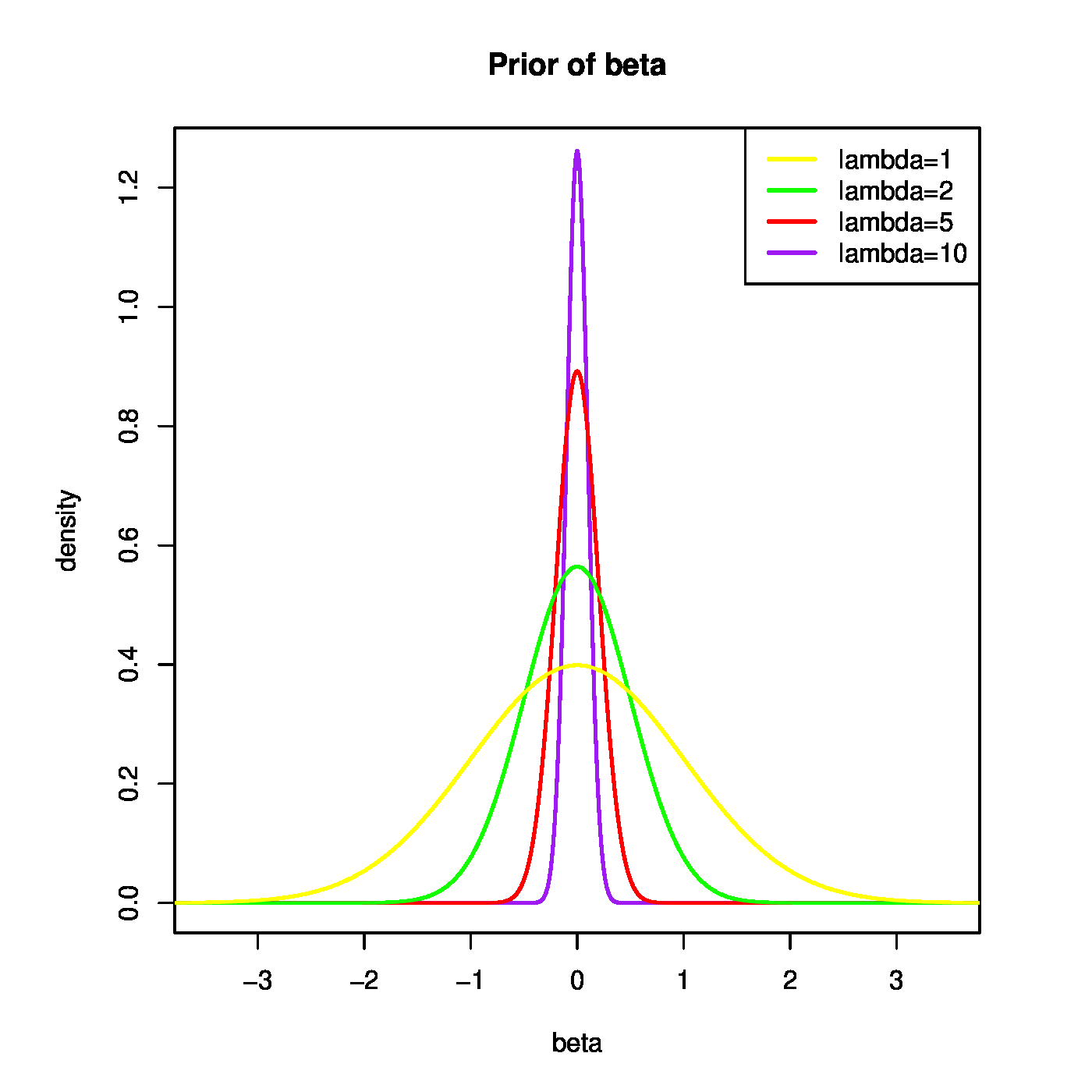
Ridge regression has a close connection to Bayesian linear regression. Bayesian linear regression assumes the parameters [math]\bbeta[/math] and [math]\sigma^2[/math] to be the random variables, while at the same time considering [math]\mathbf{X}[/math] and [math]\mathbf{Y}[/math] as fixed. Within the regression context, the commonly chosen priors of [math]\bbeta[/math] and [math]\sigma^2[/math] are [math]\bbeta \, | \, \sigma^2 \sim \mathcal{N}(\mathbf{0}_p, \sigma^2 \lambda^{-1} \mathbf{I}_{pp})[/math] and [math]\sigma^2 \sim \mathcal{IG}(\alpha_0, \beta_0)[/math], where [math]\mathcal{IG}[/math] denotes the inverse Gamma distribution with shape parameter [math]\alpha_0[/math] and scale parameter [math]\beta_0[/math]. The penalty parameter can be interpreted as the (scaled) precision of the prior of [math]\beta[/math], determining how informative the prior should be. A smaller penalty (i.e. precision) corresponds to a wider prior, and a larger penalty to a more informative, concentrated prior (Figure).
The joint posterior distribution of [math]\bbeta[/math] and [math]\sigma^2[/math] is, under the assumptions of the (likelihood of) the linear regression model and the priors above:
The posterior distribution can be expressed as a multivariate normal distribution. Hereto group the terms from the exponential functions that involve [math]\bbeta[/math] and manipulate as follows:
Using this result, the posterior distribution can be rewritten to:
with
Clearly, after having recognized the form of a multivariate normal density in the expression of the preceeding display, the conditional posterior mean of [math]\bbeta[/math] is [math]\mathbb{E}(\bbeta \, | \, \sigma^2, \mathbf{Y}, \mathbf{X}) = \hat{\bbeta}(\lambda)[/math]. Hence, the ridge regression estimator can be viewed as the Bayesian posterior mean estimator of [math]\bbeta[/math] when imposing a Gaussian prior on the regression parameter.
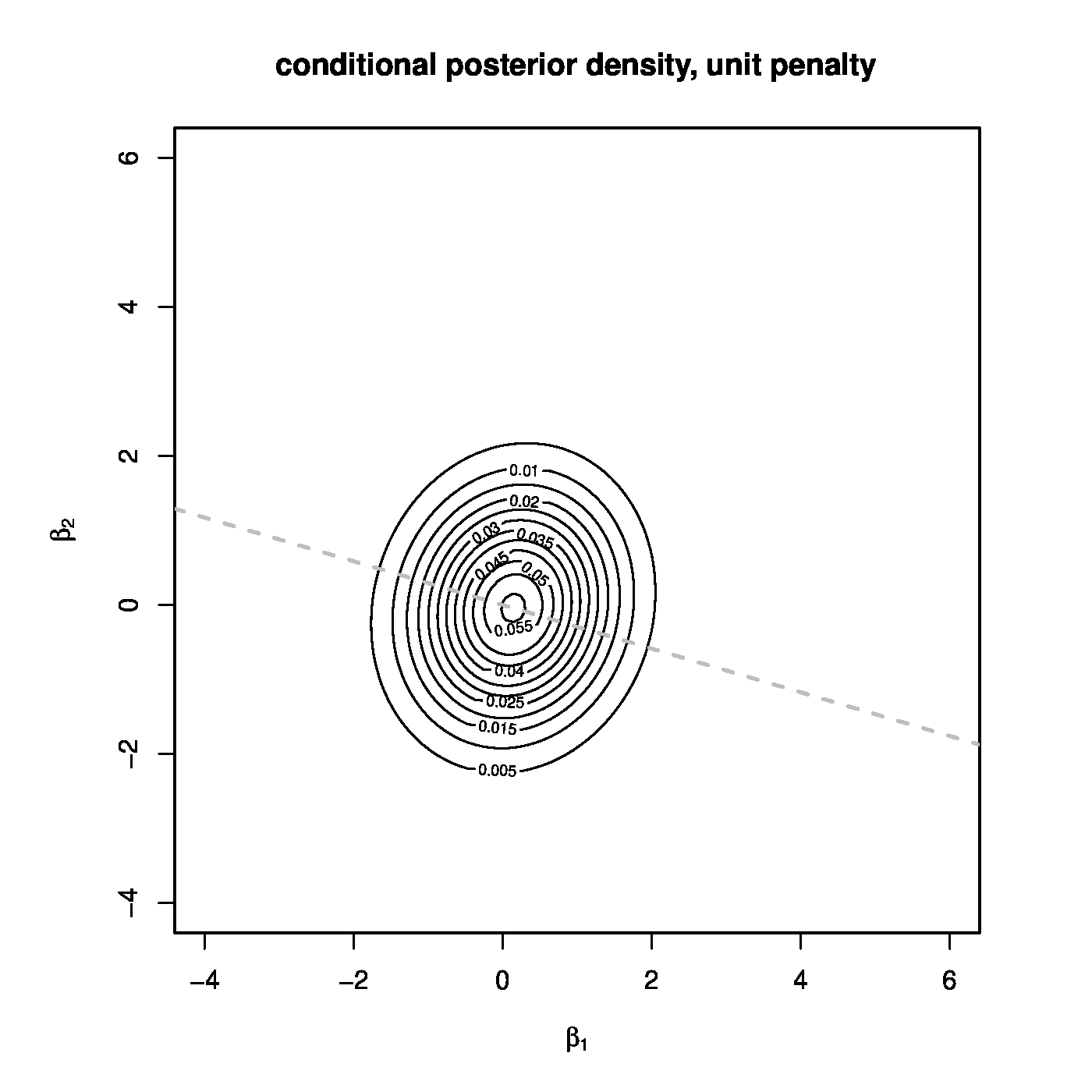
The conditional posterior distribution of [math]\bbeta[/math], [math]g_{\bbeta} (\bbeta \, | \, \sigma^2, \mathbf{Y}, \mathbf{X})[/math], related to a high-dimensional situation ([math]n=1[/math], [math]p=2[/math]) is depicted in Figure for two choices of the penalty parameter [math]\lambda[/math]. Put differently, for two different priors. In one, the left panel, [math]\lambda[/math] is arbitrarily set equal to one. The choice of the employed [math]\lambda[/math], i.e. [math]\lambda=1[/math], is irrelevant, the resulting posterior distribution only serves as a reference. This choice results in a posterior distribution that is concentrated around the Bayesian point estimate, which coincides with the ridge regression estimate. The almost spherical level sets around the point estimate may be interpreted as credible intervals for [math]\bbeta[/math]. The grey dashed line, spanned by the row of the design matrix, represents the support of the degenerated normal distribution of the frequentist's ridge regression estimate (cf. end of Section Variance ). The contrast between the degenerated frequentist and well-defined Bayesian normal distribution illustrates that -- for a suitable choice of the prior -- within the Bayesian context high-dimensionality need not be an issue with respect to the evaluation of the posterior. In the right panel of Figure the penalty parameter [math]\lambda[/math] is set equal to a very small value, i.e. [math]0 \lt \lambda \ll 1[/math]. This represents a very imprecise or uninformative prior. It results in a posterior distribution with level sets that are far from spherical and are very streched in the direction orthogonal to the subspace spanned by the row of the design matrix and indicates in which direction there is most uncertainty with respect to [math]\bbeta[/math]. In particular, when [math]\lambda \downarrow 0[/math], the level sets loose their ellipsoid form and the posterior collapses to the degenerated normal distribution of the frequentist's ridge regression estimate. Finally, in combination with the left-hand plot this illustrates the large effect of the prior in the high-dimensional context.
With little extra work we may also obtain the conditional posterior of [math]\sigma^2[/math] from the joint posterior distribution:
in which one can recognize the shape of an inverse gamma distribution. The relevance of this conditional distribution will be made clear in Section Markov chain Monte Carlo .
The Bayesian point estimator minimizes the Bayes risk, over a prior distribution. The risk cannot be used to choose the prior c.q. the penalty parameter [math]\lambda[/math]. This can be seen from the risk of the Bayesian regression estimator. The Bayes risk of the Bayesian regression estimator over the normal prior [math]\bbeta \sim \mathcal{N}(\mathbf{0}_p, \sigma^2 \lambda^{-1} \mathbf{I}_{pp})[/math] is:
in which [math]\mathbf{W}_{\lambda} = (\mathbf{X}^{\top} \mathbf{X} + \lambda \mathbf{I}_{pp})^{-1} \mathbf{X}^{\top} \mathbf{X}[/math] and we have used i) the previously derived explicit expression of the ridge estimator's MSE, ii) the expectation of the quadratic form of a multivariate random variable [2], iii) the singular value decomposition of [math]\mathbf{X}[/math] with singular values [math]d_{jj}[/math], and iv) the fact that the trace of a square matrix equals the sum of its eigenvalues. As the ridge estimator coincides with the posterior mean, this is the minimal achievable MSE under a zero-centered normal prior with an uncorrelated and equivariant covariance matrix.
One may now be tempted to choose the [math]\lambda[/math] that minimizes the Bayes risk as optimal and use it in the evaluation of the ridge regression estimator. However, above the Bayes risk of the ridge estimator factorizes with respect to [math]\sigma^2[/math] and [math]\lambda[/math]. Hence, the larger the hyperparameter [math]\lambda[/math] the lower the Bayes risk of the ridge estimator. In particular, its Bayes risk converges to zero as [math]\lambda \rightarrow \infty[/math]. This can be understood as follows. The limit corresponds to an infinite precision of the prior, thus reducing the contribution of the MSE's variance term to zero. Moreover, as the ridge estimator shrinks towards zero and the prior distribution of [math]\bbeta[/math] has a zero mean, the bias too vanishes as [math]\lambda \rightarrow \infty[/math]. In particular, the [math]\lambda[/math] that minimizes the Bayes risk is thus a useless suggestion for the ridge regression estimator. The failure to produce a useful choice of the penalty (or hyper-) paramer is due to the fact that the Bayes estimator is defined with respect to a specific prior, i.e. corresponding to a specific value of [math]\lambda[/math], and not a class of priors relating to all [math]\lambda \in \mathbb{R}_{\gt0}[/math].
The calculation of the Bayes risk above relates the Bayesian and frequentist statements on the MSE of the ridge estimator. For the latter revisit Theorem of Section Mean squared error , which warrants the existence of a [math]\lambda[/math] such that the resulting ridge estimator has a superior MSE over that of the ML estimator. This result made no assumption on (the distribution of) [math]\bbeta[/math]. In fact, it can be viewed as a statement of the MSE conditional on [math]\bbeta[/math]. The Bayesian result integrates out the uncertainty -- specified by the prior -- in [math]\bbeta[/math] from the (frequentist's) conditional MSE to arrive at the unconditional MSE.
In general, any penalized estimator has a Bayesian equivalent. That is generally not the posterior mean as for the ridge regression estimator, which is more a coincide due to the normal form of its likelihood and conjugate prior. A penalized estimator always coincides with the Bayesian MAP (Mode A Posteriori) estimator. Then, the MAP estimates estimates a parameter [math]\ttheta[/math] by the mode, i.e. the maximum, of the posterior density. To see the equivalence of both estimators we derive the MAP estimator. The latter is defined as:
To find the maximum of the posterior density take the logarithm (which does not change the location of the maximum) and drop terms that do not involve [math]\ttheta[/math]. The MAP estimator then is:
The first summand on the right-hand side is the loglikelihood, while the second is the logarithm of the prior density. The latter, in case of a normal prior, is proportional to [math]\sigma^{-2} \lambda \| \bbeta \|_2^2[/math]. This is -- up to some factors -- exactly the loss function of the ridge regression estimator. If the quadratic ridge penalty is replaced by different one, it is easy to see what form the prior density should have in order for both estimators -- the penalized and MAP -- to coincide.
Markov chain Monte Carlo
The ridge regression estimator was shown to coincide with the posterior mean of the regression parameter when it is endowed with a normal prior. This particular choice of the prior yielded a closed form expression of the posterior, and its mean. Prior distributions that result in well-characterized posterior ones are referred to as conjugate. E.g., for the standard linear regression model a normal prior is conjugate. Conjugate priors, however, are the exception rather than the rule. In general, an arbitrary prior distribution will not result in a posterior distribution that is familiar (amongst others due to the fact that its normalizing constant is not analytically evaluable). For instance, would a wildly different prior for the regression parameter be chosen, its posterior is unlikely to be analytically known. Then, although analytically unknown, such posteriors can be investigated in silico by means of the Markov chain Monte Carlo (MCMC) method. In this subsection the MCMC method is explained and illustrated -- despite its conjugancy -- on the linear regression model with a normal prior.
Markov chain Monte Carlo is a general purpose technique for sampling from complex probabilistic models/distributions. It draws a sample from a Markov chain that has been constructed such that its stationary distribution equals the desired distribution (read: the analytically untractable posterior distribution). The chain is, after an initial number of iterations (called the burn-in period), believed to have converged and reached stationarity. A sample from the stationary chain is then representative of the desired distribution. Statistics derived from this sample can be used to characterize the desired distribution. Hence, estimation is reduced to setting up and running a Markov process on a computer.
Recall Monte Carlo integration. Assume the analytical evaluation of [math]\mathbb{E}[h(\mathbf{Y})] = \int h(\mathbf{y}) \pi(\mathbf{y}) d \mathbf{y}[/math] is impossible. This quantity can nonetheless be estimated when samples from [math]\pi(\cdot)[/math] can be drawn. For, if [math]\mathbf{Y}^{(1)}, \ldots, \mathbf{Y}^{(T)} \sim \pi(\mathbf{y})[/math], the integral may be estimated by: [math]\widehat{\mathbb{E}}[h(\mathbf{Y})] = T^{-1} \sum_{t=1}^T h(\mathbf{Y}^{(t)})[/math]. By the law of large numbers this is a consistent estimator, i.e. if [math]T \rightarrow \infty[/math] the estimator converges (in probability) to its true value. Thus, irrespective of the specifics of the posterior distribution [math]\pi(\cdot)[/math], if one is able to sample from it, its moments (or other quantities) can be estimated. Or, by the same principle it may be used in the Bayesian regression framework of Section Relation to ridge regression to sample from the marginal posterior of [math]\bbeta[/math],
given that it of [math]\sigma^2[/math] is known. By drawing a sample [math]\sigma^{2,(1)}, \ldots, \sigma^{2,(T)}[/math] of [math]g_{\sigma^2}(\sigma^2 \, | \, \mathbf{Y}, \mathbf{X}) [/math] and, subsequently, a sample [math]\{ \bbeta^{(t)} \}_{t=1}^T[/math] from [math]g_{\bbeta}( \bbeta \, | \, \sigma^{2,(t)}, \mathbf{Y}, \mathbf{X})[/math] for [math]t=1, \ldots, T[/math], the parameter [math]\sigma^2[/math] has effectively been integrated out and the resulting sample of [math]\bbeta^{(1)}, \bbeta^{(2)}, \ldots, \bbeta^{(T)}[/math] is from [math]f_{\bbeta}( \bbeta \, | \, \mathbf{Y}, \mathbf{X})[/math].
It is clear from the above that the ability to sample from the posterior distribution is key to the estimation of the parameters. But this sampling is hampered by knowledge of the normalizing constant of the posterior. This is overcome by the acceptance-rejection sampler. This sampler generates independent draws from a target density [math]\pi(\mathbf{y}) = f(\mathbf{y}) / K[/math], where [math]f(\mathbf{y})[/math] is an unnormalized density and [math]K[/math] its (unknown) normalizing constant. The sampler relies on the existence of a density [math]h(\mathbf{y})[/math] that dominates [math]f(\mathbf{y})[/math], i.e., [math]f(\mathbf{y}) \leq c \, h(\mathbf{y})[/math] for some known [math]c \in \mathbb{R}_{\gt0}[/math], from which it is possible to simulate. Then, in order to draw a [math]\mathbf{Y}[/math] from the posterior [math]\pi(\cdot)[/math] run Algorithm Acceptance-rejection sampling:
- densities [math]f(\cdot)[/math] and [math]h(\cdot)[/math];
- constant [math]c \gt0[/math];
- Generate [math]\mathbf{Y}[/math] from [math]h(\cdot)[/math]
- Generate [math]U[/math] from the uniform distribution [math]\mathcal{U}[0,1][/math]
- if [math]U \leq f(\mathbf{Y}) \, / \,[ c \times h(\mathbf{Y})][/math] then
- Return [math]\mathbf{Y}[/math].
- else
- Go back to line 1.
- end
A proof that this indeed yields a random sample from [math]\pi(\cdot)[/math] is given in [3]. For this method to be computationally efficient, [math]c[/math] is best set equal to [math]\sup_{\mathbf{y}} \{ f(\mathbf{y})/h(\mathbf{y}) \}[/math]. The R-script below illustrates this sampler for the unnormalized density [math]f(y) = \cos^2(2 \pi y)[/math] on the unit interval. As [math]\max_{y \in [0,1]} \cos^2(2 \pi y) = 1[/math], the density [math]f(y)[/math] is dominated by the density function of the uniform distribution [math]\mathcal{U}[0,1][/math] and, hence, the script uses [math]h(y) = 1[/math] for all [math]y \in [0, 1][/math].
# define unnormalized target density
cos2density <- function(x){ (cos(2*pi*x))^2 }
# define acceptance-rejection sampler
acSampler <- function(n){
draw <- numeric()
for(i in 1:n){
accept <- FALSE
while (!accept){
Y <- runif(1)
if (runif(1) <= cos2density(Y)){
accept <- TRUE
draw <- c(draw, Y)
}
}
}
return(draw)
}
# verify by eye-balling
hist(acSampler(100000), n=100)
In practice, it may not be possible to find a density [math]h(x)[/math] that dominates the unnormalized target density [math]f(\mathbf{y})[/math] over the whole domain. Or, the constant [math]c[/math] is too large, yielding a rather small acceptance probability, which would make the acceptance-rejection sampler impractically slow. A different sampler is then required.
The Metropolis-Hastings sampler overcomes the problems of the acceptance-rejection sampler and generates a sample from a target distribution that is known up to its normalizing constant. Hereto it constructs a Markov chain that converges to the target distribution. A Markov chain is a sequence of random variables [math]\{ \mathbf{Y}_t \}_{t=1}^{\infty}[/math] with [math]\mathbf{Y}_t \in \mathbb{R}^p[/math] for all [math]t[/math] that exihibit a simple dependence relationship among subsequent random variables in the sequence. Here that simple relationship refers to the fact/assumption that the distribution of [math]\mathbf{Y}_{t+1}[/math] only depends on [math]\mathbf{Y}_t[/math] and not on the part of the sequence preceeding it. The Markov chain's random walk is usually initiated by a single draw from some distribution, resulting in [math]\mathbf{Y}_1[/math]. From then on the evolution of the Markov chain is described by the transition kernel. The transition kernel is a the conditional density [math]g_{\mathbf{Y}_{t+1} \, | \, \mathbf{Y}_t} (\mathbf{Y}_{t+1} = \mathbf{y}_a \, | \, \mathbf{Y}_t =\mathbf{y}_b)[/math] that specifies the distribution of the random variable at the next instance given the realization of the current one. Under some conditions (akin to aperiodicity and irreducibility for Markov chains in discrete time and with a discrete state space), the influence of the initiation washes out and the random walk converges. Not to a specific value, but to a distribution. This is called the stationary distribution, denoted by [math]\varphi (\mathbf{y})[/math], and [math]\mathbf{Y}_t \sim \varphi (\mathbf{y})[/math] for large enough [math]t[/math]. The stationary distribution satisfies:
That is, the distribution of [math]\mathbf{Y}_{t+1}[/math], obtained by marginalization over [math]\mathbf{Y}_{t}[/math], coincides with that of [math]\mathbf{Y}_{t}[/math]. Put differently, the mixing by the transition kernel does not affect the distribution of individual random variables of the chain. To verify that a particular distribution [math]\varphi(\mathbf{y})[/math] is the stationary one it is sufficient to verify it satisfies the detailed balance equations:
for all choices of [math]\mathbf{y}_a, \mathbf{y}_b \in \mathbb{R}^p[/math]. If a Markov chain satisfies this detailed balance equations, it is said to be reversible. Reversibility means so much as that, from the realizations, the direction of the chain cannot be discerned as: [math]f_{\mathbf{Y}_t,\mathbf{Y}_{t+1}} (\mathbf{y}_a, \mathbf{y}_b ) = f_{\mathbf{Y}_t,\mathbf{Y}_{t+1}} (\mathbf{y}_b, \mathbf{y}_a )[/math], that is, the probability of starting in state [math]\mathbf{y}_a[/math] and finishing in state [math]\mathbf{y}_b[/math] equals that of starting in [math]\mathbf{y}_b[/math] and finishing in [math]\mathbf{y}_a[/math]. The sufficiency of this condition is evident after its integration on both sides with respect to [math]\mathbf{y}_b[/math], from which condition (\ref{form.stationary.dist.markov.chain}) follows.
MCMC assumes the stationary distribution of the Markov chain to be known up to a scalar -- this is the target density from which is to be sampled -- but the transition kernel is unknown. This poses a problem as the transition kernel is required to produce a sample from the target density. An arbitrary kernel is unlikely to satisfy the detailed balance equations with the desired distribution as its stationary one. If indeed it does not, then:
for some [math]\mathbf{y}_a[/math] and [math]\mathbf{y}_b[/math]. This may (loosely) be interpreted as that the process moves from [math]\mathbf{y}_a[/math] to [math]\mathbf{y}_b[/math] too often and from [math]\mathbf{y}_b[/math] to [math]\mathbf{y}_a[/math] too rarely. To correct this the probability of moving from [math]\mathbf{y}_b[/math] to [math]\mathbf{y}_b[/math] is introduced to reduce the number of moves from [math]\mathbf{y}_a[/math] to [math]\mathbf{y}_b[/math]. As a consequence not always a new value is generated, the algorithm may decide to stay in [math]\mathbf{y}_a[/math] (as opposed to acceptance-rejection sampling). To this end a kernel is constructed and comprises compound transitions that propose a possible next state which is simultaneously judged to be acceptable or not. Hereto take an arbitrary candidate-generating density function [math]g_{\mathbf{Y}_{t+1} \, | \, \mathbf{Y}_t} (\mathbf{y}_{t+1} \, | \, \mathbf{y}_{t})[/math]. Given the current state [math]\mathbf{Y}_{t} = \mathbf{y}_{t}[/math], this kernel produces a suggestion [math]\mathbf{y}_{t+1}[/math] for the next point in the Markov chain. This suggestion may take the random walk too far afield from the desired and known stationary distribution. If so, the point is to be rejected in favour of the current state, until a new suggestion is produced that is satisfactory close to or representative of the stationary distribution. Let the probability of an acceptable suggestion [math]\mathbf{y}_{t+1}[/math], given the current state [math]\mathbf{Y}_t = \mathbf{y}_t[/math], be denoted by [math]\mathcal{A}(\mathbf{y}_{t+1} \, | \, \mathbf{y}_t)[/math]. The thus constructed transition kernel then formalizes to:
where [math]\mathcal{A} (\mathbf{y}_{a} \, | \, \mathbf{y_b})[/math] is the acceptance probability of the suggestion [math]\mathbf{y}_a[/math] given the current state [math]\mathbf{y}_b[/math] and is defined as:
Furthermore, [math]\delta_{\mathbf{y_b}} (\cdot)[/math] is the Dirac delta function and [math]r(\mathbf{y}_a) = 1 - \int_{\mathbb{R}^p} g_{\mathbf{Y}_{t+1} \, | \, \mathbf{Y}_t} (\mathbf{y}_a \, | \, \mathbf{y}_b) \mathcal{A} (\mathbf{y}_{a} \, | \, \mathbf{y_b}) \, d\mathbf{y}_a[/math], which is associated with the rejection. The transition kernel [math]h_{\mathbf{Y}_{t+1} \, | \, \mathbf{Y}_{t}}(\cdot \, | \, \cdot)[/math] equipped with the above specified acceptance probability is referred to as the Metropolis-Hastings kernel. Note that, although the normalizing constant of the stationary distribution may be unknown, the acceptance probability can be evaluated as this constant cancels out in the [math]\varphi(\mathbf{y}_{a}) \, [\varphi(\mathbf{y}_{b})]^{-1}[/math] term.
It rests now to verify that [math]\varphi(\cdot)[/math] is the stationary distribution of the thus defined Markov process. Hereto first the reversibility of the proposed kernel is assessed. The contribution of the second summand of the kernel to the detailed balance equations, [math]r(\mathbf{y}_a) \, \delta_{\mathbf{y_b}} ( \mathbf{y}_a ) \varphi ( \mathbf{y}_b)[/math], exists only if [math]\mathbf{y}_a = \mathbf{y}_b[/math]. Thus, if it contributes to the detailed balance equations, it does so equally on both sides of the equation. It is now left to assess whether:
To verify this equality use the definition of the acceptance probability from which we note that if [math]\mathcal{A} (\mathbf{y}_{a} \, | \, \mathbf{y_b}) \leq 1[/math] (and thus [math]\mathcal{A} (\mathbf{y}_{a} \, | \, \mathbf{y_b}) = [ \varphi(\mathbf{y}_{a}) g_{\mathbf{Y}_{t+1} \, | \, \mathbf{Y}_t} (\mathbf{y}_{b} \, | \, \mathbf{y}_a) ] [ \varphi(\mathbf{y}_{b}) g_{\mathbf{Y}_{t+1} \, | \, \mathbf{Y}_t} (\mathbf{y}_a \, | \, \mathbf{y}_b) ]^{-1}[/math]), then [math]\mathcal{A} (\mathbf{y}_{b} \, | \, \mathbf{y}_a) = 1[/math] (and vice versa). In either case, the equality in the preceeding display holds, and thereby, together with the observation regarding the second summand, so do the detailed balance equations for Metropolis-Hastings kernel. Then, [math]\varphi(\cdot)[/math] is indeed the stationary distribution of the constructed transition kernel [math]h_{\mathbf{Y}_{t+1} \, | \, \mathbf{Y}_t} (\mathbf{y}_a \, | \, \mathbf{y}_b)[/math] as can be verified from Condition (\ref{form.stationary.dist.markov.chain}):
in which the reversibility of [math]h_{\mathbf{Y}_{t+1} \, | \, \mathbf{Y}_t} (\cdot, \cdot)[/math], the definition of [math]r(\cdot)[/math], and the properties of the Dirac delta function have been used.
The Metropolis-Hastings algorithm is now described by:
- densities [math]f(\cdot)[/math] and [math]h(\cdot)[/math];
- constant [math]c \gt0[/math];
- Choose starting value [math]x^{(0)}[/math]
- Generate [math]y[/math] from [math]q(x^{(j)}, \cdot)[/math] and [math]U[/math] from [math]\mathcal{U}[0,1][/math]
- if [math]U \leq \alpha(X^{(j)}, y)[/math] then
- set [math]x^{(j+1)} = y[/math].
- else
- set [math]x^{(j+1)} = x^{(j)}[/math].
- end
- Return the values [math]\{ x^{(1)}, x^{(2)}, x^{(3)}, \ldots, \}[/math]
The convergence rate of this sequence is subject of on-going research.
The following R-script shows how the Metropolis-Hastings sampler can be used to from a mixture of two normals [math]f(y) = \theta \phi_{\mu=2, \sigma^2=0.5}(y) + (1-\theta) \phi_{\mu=-2, \sigma^2=2}(y)[/math] with [math]\theta = {1}{4}[/math]. The sampler assumes this distribution is known up to its normalizing constant and uses its unnormalized density [math]\exp[-(y-2)^2] + 3\exp[-(y+2)^2/4][/math]. The sampler employs the Cauchy distribution centered at the current state as transition kernel from a candidate for the next state is drawn. The chain is initiated arbitrarily with [math]Y^{(1)} = 1[/math].
# define unnormalized mixture of two normals
mixDensity <- function(x){
# unnormalized mixture of two normals
exp(-(x-2)^2) + 3*exp(-(x+2)^2/4)
}
# define the MCMC sampler
mcmcSampler <- function(n, initDraw){
draw <- initDraw
for (i in 1:(n-1)){
# sample a candidate
candidate <- rcauchy(1, draw[i])
# calculate acceptance probability:
alpha <- min(1, mixDensity(candidate) / mixDensity(draw[i]) *
dcauchy(draw[i], candidate) / dcauchy(candidate, draw[i]))
# accept/reject candidate
if (runif(1) < alpha){
draw <- c(draw, candidate)
} else {
draw <- c(draw, draw[i])
}
}
return(draw)
}
# verify by eye-balling
Y <- mcmcSampler(100000, 1)
hist(Y[-c(1:1000)], n=100)
The histogram (not shown) shows that the sampler, although using a unimodal kernel, yields a sample from the bimodal mixture distribution.
The Gibbs sampler is a particular version of the MCMC algorithm. The Gibbs sampler enhances convergence to the stationary distribution (i.e. the posterior distribution) of the Markov chain. It requires, however, the full conditionals of all random variables (here: model parameters), i.e. the conditional distributions of one random variable given all others, to be known analytically. Let the random vector [math]\mathbf{Y}[/math] for simplicity -- more refined partitions possible -- be partioned as [math](\mathbf{Y}_{a}, \mathbf{Y}_{b})[/math] with subscripts [math]a[/math] and [math]b[/math] now refering to index sets that partition the random vector [math]\mathbf{Y}[/math] (instead of the previously employed meaning of referring to two -- possibly different -- elements from the state space). The Gibbs sampler thus requires that both [math]f_{\mathbf{Y}_{a} \, | \, \mathbf{Y}_b} (\mathbf{y}_{a}, \mathbf{y}_{b})[/math] and [math]f_{\mathbf{Y}_b \, | \, \mathbf{Y}_a} (\mathbf{y}_a, \mathbf{y}_b)[/math] are known. Being a specific form of the MCMC algorithm the Gibbs sampler seeks to draw [math]\mathbf{Y}_{t+1} = (\mathbf{Y}_{a,t+1}, \mathbf{Y}_{b,t+1})[/math] given the current state [math]\mathbf{Y}_{t} = (\mathbf{Y}_{a,t}, \mathbf{Y}_{b,t})[/math]. It draws, however, only a new instance for a single element of the partition (e.g. [math]\mathbf{Y}_{a,t+1}[/math]) keeping the remainder of the partition (e.g. [math]\mathbf{Y}_{b,t}[/math]) temporarily fixed. Hereto define the transition kernel:
Using the definition of the conditional density the acceptance probability for the [math]t+1[/math]-th proposal of the subvector [math]\mathbf{Y}_a[/math] then is:
The acceptance probability of each proposal is thus one (which contributes to the enhanced convergence of the Gibbs sampler to the joint posterior). Having drawn an acceptable proposal for this element of the partition, the Gibbs sampler then draws a new instance for the next element of the partition, i.e. now [math]\mathbf{Y}_b[/math], keeping [math]\mathbf{Y}_a[/math] fixed. This process of subsequently sampling each partition element is repeated until enough samples have been drawn.
To illustrate the Gibbs sampler revisit Bayesian regression. In Section Relation to ridge regression the full conditional distributions of [math]\bbeta[/math] and [math]\sigma^2[/math] were derived. The Gibbs sampler now draws in alternating fashion from these conditional distributions (see Algorithm for its pseudo-code).
- sample size [math]T[/math]
- length of burn in period [math]T_{\mbox{{\tiny burn-in}}}[/math]
- thinning factor [math]f_{{\mbox{{\tiny thinning}}}}[/math]
- data [math]\{\mathbf{x}_i, \mathbf{y}_i \}_{i=1}^n[/math];
- conditional distributions [math]f_{\sigma^2 \, | \, \bbeta} (\sigma^2, \bbeta; \{\mathbf{x}_i, \mathbf{y}_i \}_{i=1}^n)[/math] and [math]f_{\beta \, | \, \sigma^2} (\bbeta, \sigma^2; \{\mathbf{x}_i, \mathbf{y}_i \}_{i=1}^n)[/math].
- initialize [math](\bbeta_0, \sigma^2_{0})[/math].
- for [math]t = 1[/math] to [math]T_{\mbox{{\tiny burn-in}}} + T f_{{\mbox{{\tiny thinning}}}}[/math] do
- draw [math]\bbeta_{t}[/math] from conditional distribution [math]f_{\bbeta \, | \, \sigma^2} (\bbeta, \sigma_{t-1}^2; \{\mathbf{x}_i, \mathbf{y}_i \}_{i=1}^n)[/math],
- draw [math]\sigma_{t}^2[/math] from conditional distribution [math]f_{\sigma^2 \, | \, \bbeta} (\sigma^2, \bbeta_t; \, \{\mathbf{x}_i, \mathbf{y}_i \}_{i=1}^n)[/math].
- end
- Remove the first [math]T_{\mbox{{\tiny burn-in}}}[/math] draws (representing the burn-in phase).
- Select every [math]f_{\mbox{{\tiny thinning}}}[/math]-th sample (thinning).
Empirical Bayes
Empirical Bayes (EB) is a branch of Bayesian statistics that meets the subjectivity criticism of frequentists. Instead of fully specifying the prior distribution empirical Bayesians identify only its form. The hyper parameters of this prior distribution are left unspecified and are to be found empirically. In practice, these hyper-parameters are estimated from the data at hand. However, the thus estimated hyper parameters are used to obtain the Bayes estimator of the model parameters. As such the data are then used multiple times. This is usually considered an inappropriate practice but is deemed acceptable when the number of model parameters is large in comparison to the number of hyper parameters. Then, the data are not used twice but ‘[math]1+\varepsilon[/math]’-times (i.e. once-and-a-little-bit) and only little information from the data is spent on the estimation of the hyper parameters. Having obtained an estimate of the hyper parameters, the prior is fully known, and the posterior distribution (and summary statistics thereof) are readily obtained by Bayes' formula.
The most commonly used procedure for the estimation of the hyper parameters is marginal likelihood maximization, which is a maximum likelihood-type procedure. But the likelihood cannot directly be maximized with respect to the hyper parameters as it contains the model parameters that are assumed to be random within the Bayesian framework. This may be circumvented by choosing a specific value for the model parameter but would render the resulting hyper parameter estimates dependent on this choice. Instead of maximization with the model parameter set to a particular value one would preferably maximize over all possible realizations. The latter is achieved by marginalization with respect to the random model parameter, in which the (prior) distribution of the model parameter is taken into account. This amounts to integrating out the model parameter from the posterior distribution, i.e. [math]\int P(\mathbf{Y} = \mathbf{y} \, | \, \theta) \, \pi (\theta) \, d \theta[/math], resulting in the so-called marginal posterior. After marginalization the specifics of the model parameter have been discarded and the marginal posterior is a function of the observed data and the hyper parameters. The estimator of the hyper parameter is now defined as the maximizer of this marginal posterior.
To illustrate the estimation of hyper parameters of the Bayesian linear regression model through marginal likelihood maximization assume the regression parameter [math]\bbeta[/math] and and the error variance [math]\sigma^2[/math] to be endowed with conjugate priors: [math]\bbeta \, | \, \sigma^2 \sim \mathcal{N}(\mathbf{0}_p, \sigma^2 \lambda^{-1} \mathbf{I}_{pp})[/math] and [math]\sigma^2 \sim \mathcal{IG}(\alpha_0, \beta_0)[/math]. Three hyper parameters are thus to be estimated: the shape and scale parameters, [math]\alpha_0[/math] and [math]\beta_0[/math], of the inverse gamma distribution and the [math]\lambda[/math] parameter related to the variance of the regression coefficients. Straightforward application of the outlined marginal likelihood principle does, however, not work here. The joint prior, [math]\pi(\bbeta \, | \, \sigma^2) \pi (\sigma^2)[/math], is too flexible and does not yield sensible estimates of the hyper parameters. As interest is primarily in [math]\lambda[/math], this is resolved by setting the hyper parameters of [math]\pi (\sigma^2)[/math] such that the resulting prior is uninformative, i.e. as objectively as possible. This is operationalized as a very flat distribution. Then, with the particular choices of [math]\alpha_0[/math] and [math]\beta_0[/math] that produce an uninformative prior for [math]\sigma^2[/math], the empirical Bayes estimate of [math]\lambda[/math] is:
where the factors not involving [math]\lambda[/math] have been dropped throughout and [math]b_{1} = \beta_0 + \tfrac{1}{2} [ \mathbf{Y}^{\top} \mathbf{Y} - \mathbf{Y}^{\top} \mathbf{X} (\mathbf{X}^{\top} \mathbf{X} + \lambda \mathbf{I}_{pp} )^{-1} \mathbf{X}^{\top} \mathbf{Y} ][/math]. Prior to the maximization of the marginal likelihood the logarithm is taken. That changes the maximum, but not its location, and yields an expression that is simpler to maximize. With the empirical Bayes estimate [math]\hat{\lambda}_{eb}[/math] at hand, the Bayesian estimate of the regression parameter [math]\bbeta[/math] is [math]\hat{\bbeta}_{eb} = (\mathbf{X}^{\top} \mathbf{X} + \hat{\lambda}_{eb} \mathbf{I}_{pp})^{-1} \mathbf{X}^{\top} \mathbf{Y}[/math]. Finally, the particular choice of the hyper parameters of the prior on [math]\sigma^2[/math] is not too relevant. Most values of [math]\alpha_0[/math] and [math]\beta_0[/math] that correspond to a rather flat inverse gamma distribution yield resulting point estimates [math]\hat{\bbeta}_{eb}[/math] that do not differ too much numerically.
Conclusion
Bayesian regression was introduced and shown to be closely connected to ridge regression. Under a conjugate Gaussian prior on the regression parameter the Bayesian regression estimator coincides with the ridge regression estimator, which endows the ridge penalty with the interpretation of this prior. While an analytic expression of these estimators is available, a substantial part of this chapter was dedicated to evaluation of the estimator through resampling. The use of this resampling will be evident when other penalties and non-conjugate priors will be studied (cf. Exercise \ref{} and Sections The Bayesian connection and The Bayesian connection ). Finally, another informative procedure, empirical Bayes, to choose the penalty parameter was presented.
General References
van Wieringen, Wessel N. (2021). "Lecture notes on ridge regression". arXiv:1509.09169 [stat.ME].
References
- Bijma, F., Jonker, M. A., and van der Vaart, A. W. (2017).An Introduction to Mathematical Statistics.Amsterdam University Press
- Mathai, A. M. and Provost, S. B. (1992).Quadratic Forms in Random Variables: Theory and Applications.Dekker
- Flury, B. D. (1990).Acceptance--rejection sampling made easy.SIAM Review, 32(3), 474--476